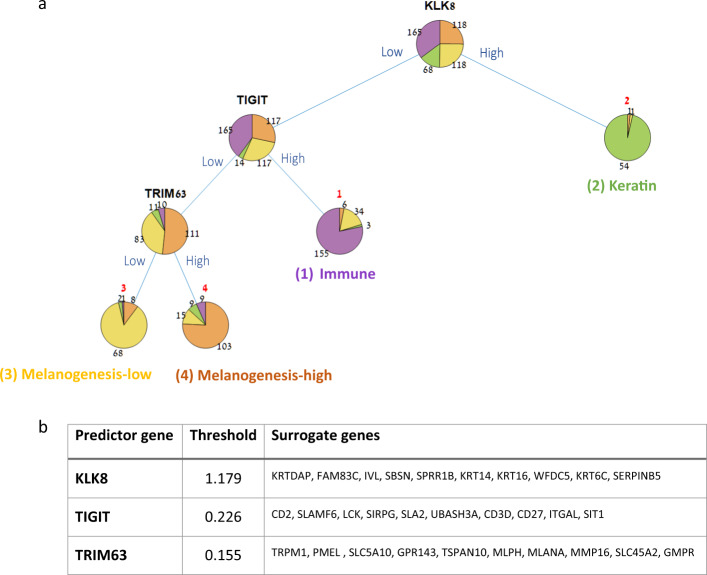
Fig. 4. A three-gene decision tree for classifying melanoma samples.
a The tree trained on the 469 TCGA samples. Classification of a new sample into one of the four subtypes is done by traversing the tree from its root to one of its leaves (representing an assignment to a subtype). Three biomarkers (shown in black) are used to determine the route along the tree: overexpression of KLK8 distinguishes the “Keratin” subtype, overexpression of TIGIT distinguishes the “Immune” subtype, and finally, overexpression of TRIM63 distinguishes the “Melanogenesis-high” from the “Melanogenesis-low” subtype. b Threshold values and surrogate genes for the three decision tree predictors as identified by the algorithm. Threshold values are used to distinguish between high and low values (based on normalized expression values), and surrogates can be used as alternatives for the respective predictor gene.