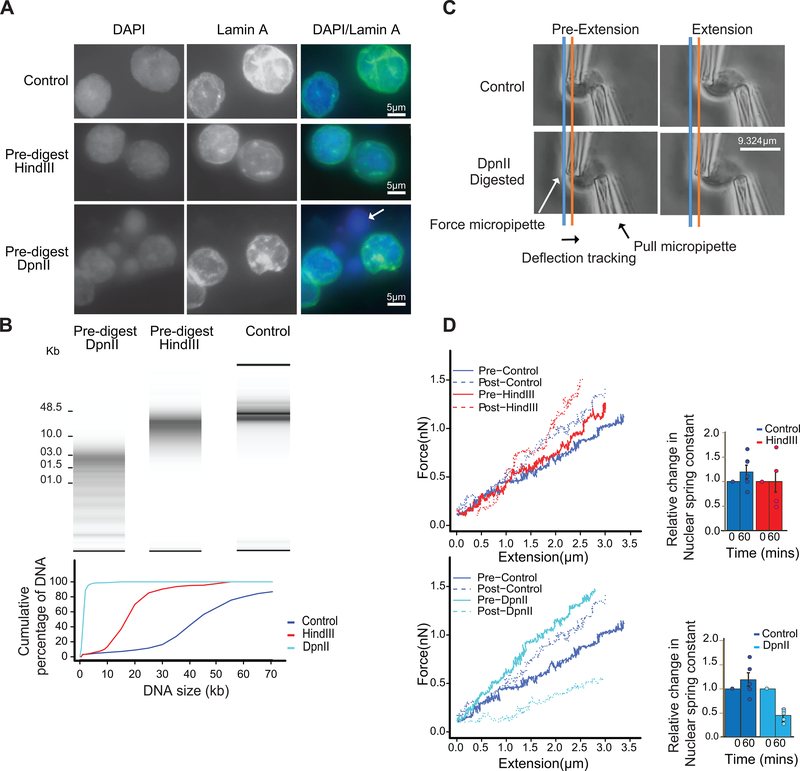
Fig. 2: Extensive fragmentation of chromatin leads to liquefied chromatin.
A: Nuclear and chromatin morphology before and after chromatin fragmentation. Top row: control nuclei in restriction buffer, middle row nuclei digested for 4 hours with HindIII. Bottom row: nuclei digested for 4 hours with DpnII. Nuclei were stained with DAPI (left column), with antibodies against Lamin A (middle column). The right column shows the overlay of the DAPI and Lamin A stained images. HindIII digestion did not lead to major alteration in nuclear morphology and chromatin appearance, while DpnII digestion led to the appearance of DAPI stained droplets (arrow) exiting the nuclei. Representative images are shown, experiment repeated twice, with dozen nuclei images per experiment.
B: Top: DNA purified from undigested nuclei, and nuclei pre-digested with DpnII and HindIII was run on a Fragment Analyzer. Bottom: cumulative DNA length distributions calculated from the Fragment Analyzer data. Representative data are shown for 1 out of 2 replicates.
C: Micromanipulation of single nuclei. Isolated nuclei were attached to two micropipettes at opposite ends. Nuclei were extended by moving the right micropipette (Extension micropipette) and the force required was calculated from the deflection of the calibrated “force” (left) pipette. Blue and orange lines indicate the position of the force pipette before and after extension for control nuclei. After digestion of nuclei with DpnII (bottom) extension required less force as indicated by the much smaller deflection of the force pipette as compared to control nuclei (see also Supplementary Movies 1 and 2).
D. Force-extension plots (left) for control nuclei before and 60 minutes after incubation in restriction buffer (pre- and post-control), for nuclei before and after digestion with DpnII, and for nuclei before and after HindIII digestion. Right panel: relative change in nuclear spring constants, calculated from the slopes of the force-extension plots shown on the left. Bars indicate standard error of the mean (n = 5 DpnII pre-digested nuclei, and n = 4 HindIII pre-digested nuclei). Bars show the average of all nuclei, dots indicate values obtained with individual nuclei.