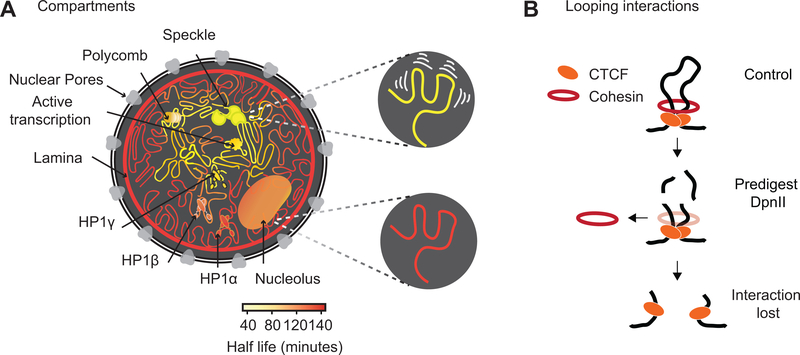
Fig. 7: Illustration of chromatin interaction dynamics in the nucleus and model for cohesin loss after chromatin digestion.
(A) Left: Schematic representation of varying chromatin interactions dynamics at different sub-nuclear domains. Shortest half-life reflects the least stable interactions (yellow), while longest half-life reflects the most stable interactions (dark orange). Nuclear subdomains differ greatly in their stability. Top right: Chromatin anchored at speckles is driven by the most dynamic interactions. Bottom right: Chromatin anchored at the nuclear lamina involves the most stable interactions.
(B) Model for how cohesin rings stabilize CTCF-CTCF loops by encircling loop bases. Top: Cohesin ring encircles loop bases at convergent CTCF sites. Middle: Pre-digestion with DpnII cuts loop into chromatin fragments <6 kb, and the cohesin ring can slide off nearby ends. Bottom: CTCF remains bound to digested chromatin fragments but interactions between CTCF-bound sites are lost.