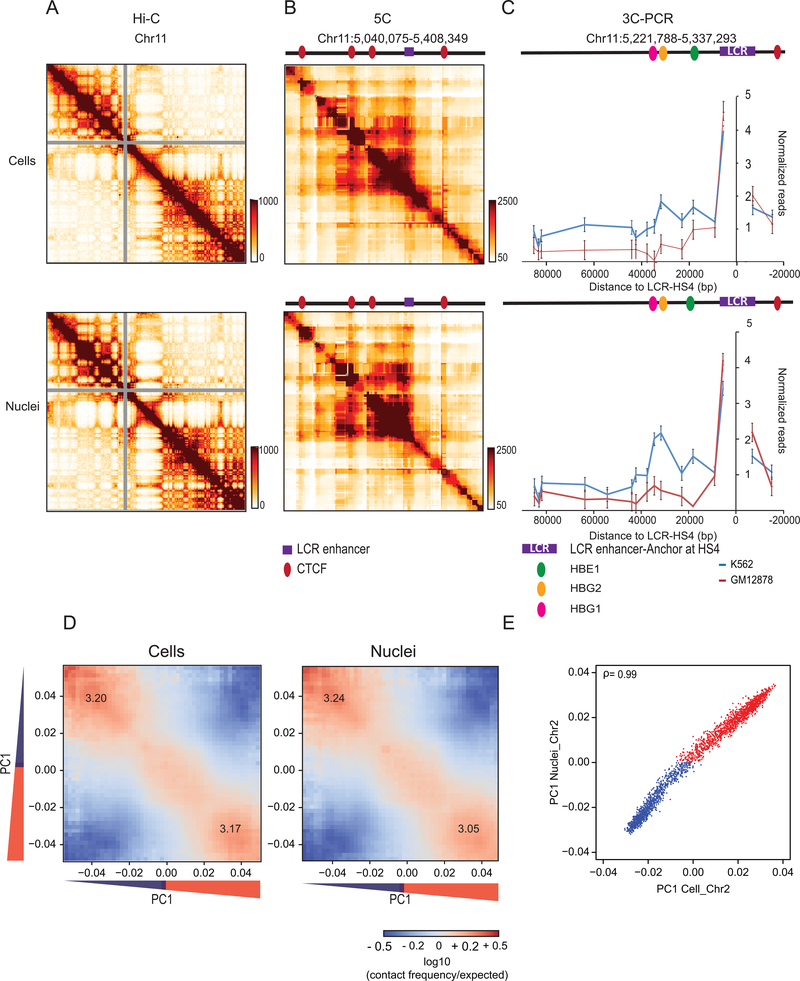
Extended Data Fig. 1: Chromosome conformation in isolated nuclei.
(A) Hi-C 2.0 intra-chromosomal interaction maps for K562 cells (top) and purified nuclei (bottom).
(B) 5C interaction map of 1 Mb region surrounding the beta-globin locus in K562 cells. Top: cells. Bottom: purified nuclei. CTCF-mediated interactions are preserved in purified nuclei. Red circles: positions of CTCF sites, purple square Beta-globin locus control region (LCR).
(C) Representative 3C-PCR (out of two experiments) for a 44,120 kb region surrounding the beta-globin LCR on chromosome 11, detects at high resolution the known looping interactions between the LCR and the expressed gamma-globin genes (HBE1, HBG2) in K562 cells. Looping interactions are not detected in GM12878 cells that do not express these genes. Top: cells. Bottom: purified nuclei. Each data point is the average of 3 PCR reactions; error bars indicate standard error of the mean.
(D) Compartmentalization saddle plots: average intra-chromosomal interaction frequencies between 100 kb bins, normalized by genomic distance. Bins are sorted by their PC1 value derived from Hi-C data obtained with K562 cells. In these plots preferential B-B interactions are in the upper left corner, and preferential A-A interactions are in the lower right corner. Numbers in the corners represent the strength of AA interactions as compared to AB interactions and BB interactions over BA interactions. Left: cells. Right: purified nuclei.
(E) Spearman correlation of PC1 in cells vs PC1 in nuclei for chromosome 2 at 100kb resolution (ρ= 0.99).