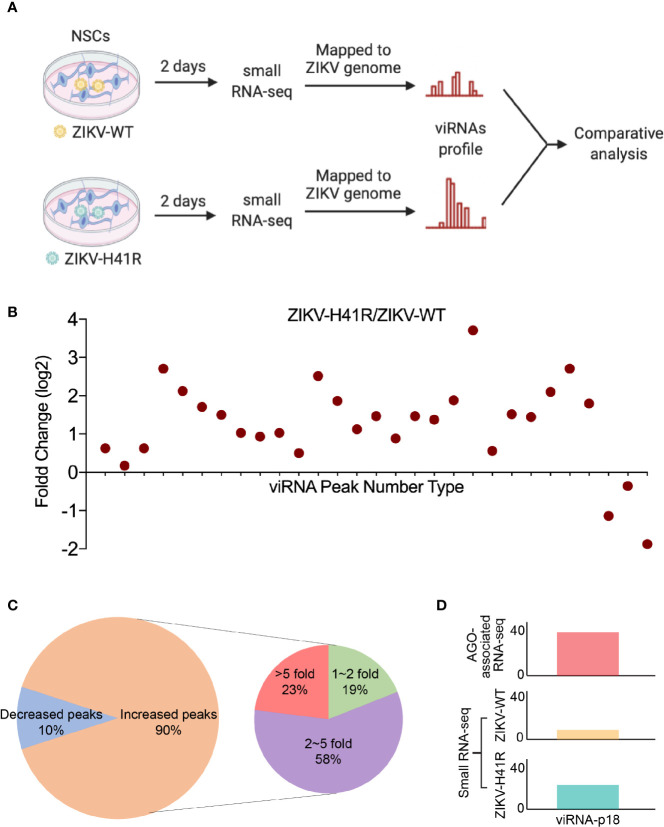
Figure 3.
The identified viRNAs are potentially the products of Dicer activity via directly deep small RNA-seq of ZIKV-infected NSCs. (A) Schematic diagram of comparative analysis for directly deep small RNA-seq in ZIKV-WT- or ZIKV-H41R-infected NSCs. (B) Comparison of read counts in 29 ZIKV viRNA peaks in ZIKV-WT- or ZIKV-H41R-infected NSCs at MOI of 1 for 2 days. The fold change of presented peaks was log2 scale. (C) 90% viRNA peaks identified from AGO-associated RNA sequencing are increased in ZIKV-H41R infected NSCs compared to ZIKV-WT infected ones. (D) Peak landscape of representative viRNA-p18 in AGO-associated RNA-seq and deep small RNA-seq. Value in the small RNA-seq (lower panel) in y-axis indicates reads per 10 million sequenced reads.