Figure 5.
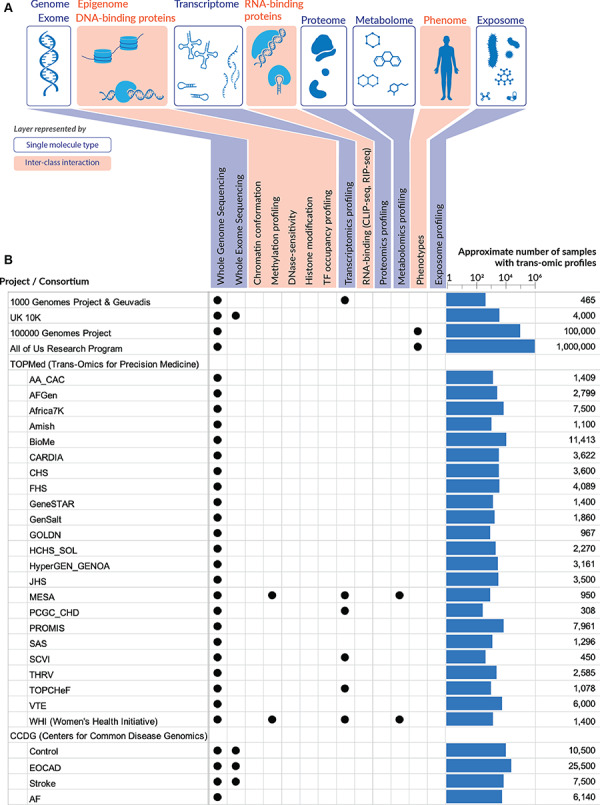
Multi-omics data. (A) The multiple layers of omics data that are now accessible to researchers. Genome/exome, transcriptome, proteome, metabolome, as well as the microbiome and chemical compounds in the exposome can be profiled by assays on a single class of molecules (DNA, RNA, proteins or small molecules), while the other layers depend on the ability to capture DNA–protein or RNA–protein interactions. The phenome is less well defined as phenotypic measures vary greatly from physical measurements to laboratory tests, from descriptive to quantitative traits. Sources of comprehensive phenotypic data comparable to the other omics can be obtained, for example, from EHRs. Beyond the genome, omics datasets become highly complicated, due to the variation across tissues and cell types. (B) Large omics datasets that are (or will be) available for CVD research. For each dataset, the number of samples being assayed across multiple omics are indicated on the right. This number is often smaller than the total number of samples/participants in a given project, because not every sample is run on multiple assays. Sources are provided in Supplementary Data.
