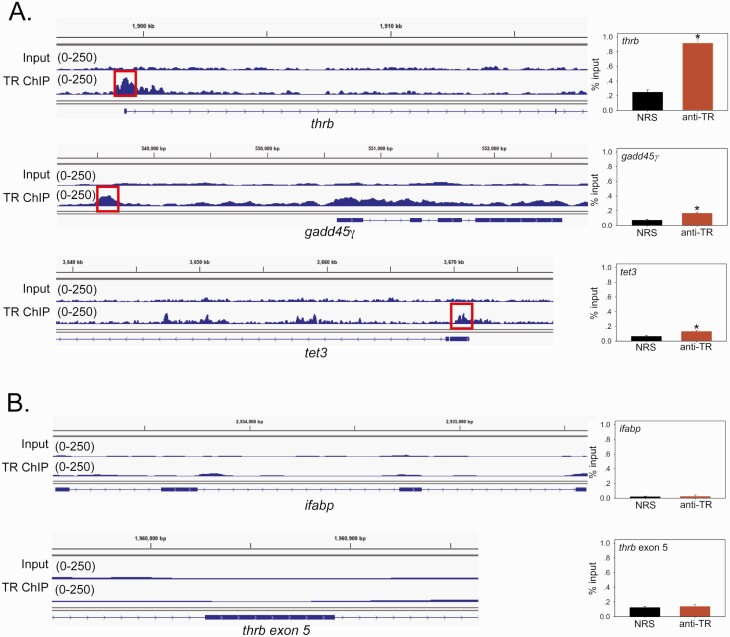
Figure 4.
Thyroid hormone receptors (TRs) associate in chromatin in metamorphic climax stage tadpole brain at genes that encode DNA demethylation enzymes. Shown are integrative genome viewer (IGV) genome browser tracks for TR ChIP-seq reads mapped to the X. tropicalis genome. We conducted a TR ChIP-seq experiment on chromatin isolated from the region of the preoptic area/thalamus/hypothalamus of metamorphic climax stage (NF stage 62) X. tropicalis tadpole brain. The input tracks are shown above the TR ChIP-seq tracks. Numbers in parentheses represent the scale for peak height. The gene structures are shown below the genome traces: lines and black filled bars represent introns and exons, respectively, and arrows indicate the direction 5’ → 3’. To the right of each genome track is a graph of a TR ChIP assay using normal rabbit serum (NRS; negative control) or rabbit antiserum to Xenopus TRs (anti-TR) that targeted the region covered by the TR ChIP-seq peak shown on the IGV genome browser track (indicated by the red box). The bars represent the mean ± SEM (n = 4). Asterisks indicate statistically significant differences between NRS and anti-TR serum (P < 0.05, Student’s independent t-test). A: IGV genome browser tracks and targeted TR ChIP assays at the thrb (XLOC_017285), gadd45γ (XLOC_017078.1), and tet3 (XLOC_025924) genes. The thrb gene, which has a TRE in its 5’ UTR (71, 72, 80), served as a positive control. The putative TRE region at gadd45γ is located 1.8 kb upstream of the TSS, and at tet3 it is in the 5’ UTR. B: IGV genome browser tracks and targeted TR ChIP assays at two negative control regions, the ifabp (XLOC_43086.1) gene and thrb exon 5.