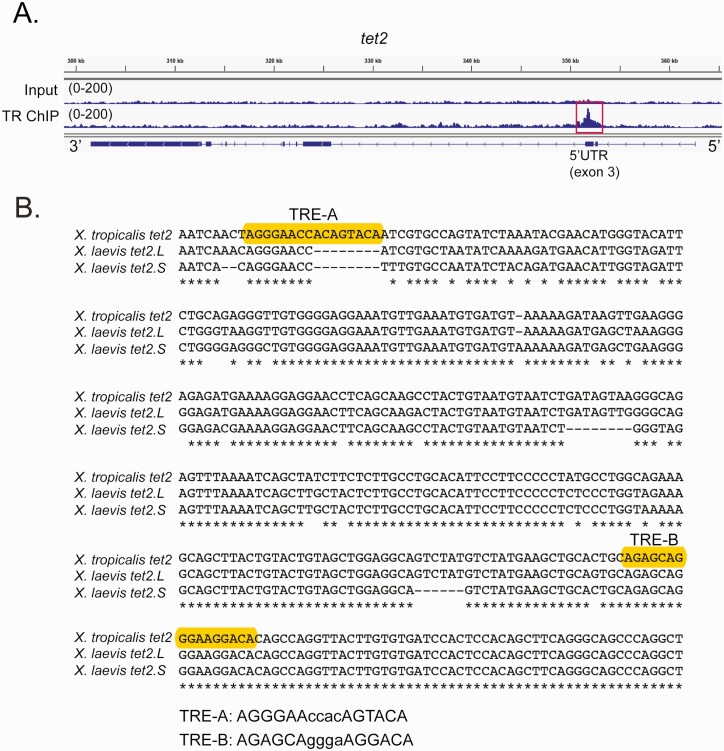
Figure 5.
Identification of putative TREs within the X. tropicalis tet2 gene. A: Genome browser tracks showing the location of a TR peak (red box) within the 5’ UTR (exon 3) of X. tropicalis tet2 (XLOC_002313) identified by TR ChIP-sequencing (see Supplemental Methods) (62) conducted on chromatin isolated from the whole brain of metamorphic climax stage (NF stage 62) tadpoles. Shown are genome tracks obtained with input and TR ChIP samples (numbers in parentheses indicate the peak height range). In the schematic under the genome browser tracks, the black boxes represent exons and the lines represent introns, and arrows on the gene indicate the orientation of the gene. B: Alignment of DNA sequences (5’ → 3’) within the third exons of X. tropicalis (358 bp) and X. laevis tet2 genes (located on chromosome 1 in both species; for X. laevis, which is pseudotetraploid, L – long chromosome, S – short chromosome). Accession numbers: X. tropicalis tet2 XM_012955515.3; X. laevis tet2.L LOC108710731; and X. laevis tet2.S LOC108706731. Shown are the locations of two predicted DR+4 TREs in X. tropicalis (only TRE-B is found in both species) identified using the sequence analysis program NHR-scan.