Figure 1.
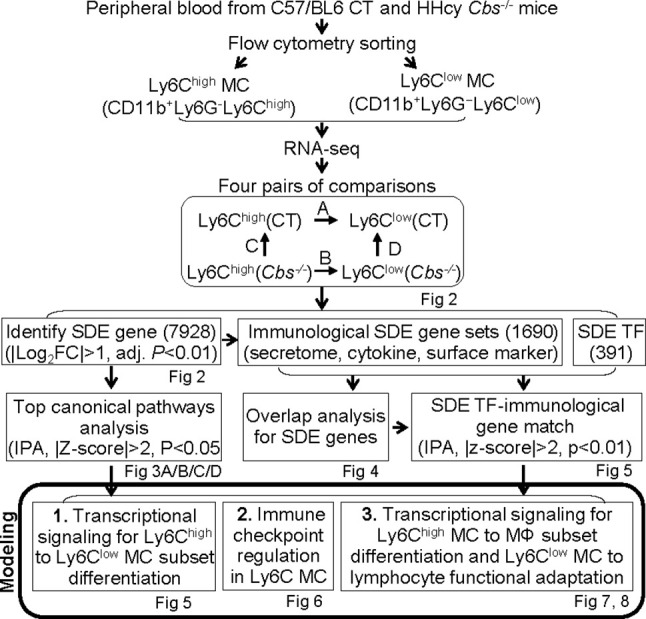
Overall strategy of the identification of Ly6C MC regulatory genes and molecule mechanism for Ly6C monocyte subset differentiation in control and Cbs -/- mice. RNA-seq were performed in Ly6Chigh (CD11b+Ly6G−Ly6Chigh) and Ly6Clow (CD11b+Ly6G−Ly6Clow) MC isolated by flow cytometry sorting from peripheral blood of C57/BL6 control and Cbs -/- mice. Transcriptome data were analyzed by performing four pairs of comparisons; (A) Ly6Chigh vs. Ly6Clow (CT), (B) Ly6Chigh vs. Ly6Clow (Cbs-/-), (C) Cbs-/- vs. CT (Ly6Chigh), (D) Cbs-/- vs. CT (Ly6Clow). We identified 7928 SDE genes using the Bioconductor suite of packages in RStudio software with the criteria of |Log2FC| more than 1 (2-FC) and adjusted P value less than 0.01. Top ingenuity pathways were identified by top-down analysis using IPA with |Z-score|>2, P value<0.05. Immunological SDE gene sets, including secretome, cytokine and surface marker were overlapped analysis and matched with corresponding upstream SDE TF by IPA upstream analysis. Three molecular signaling model system were developed, 1) Transcriptional regulation for Ly6Chigh to Ly6Clow MC subset differentiation, 2) Immune checkpoint regulation in Ly6C MC. 3) Transcriptional signaling for Ly6Chigh MC to MΦ subset differentiation and Ly6Clow MC to lymphocyte functional adaptation, CT, control, HHcy, Hyperhomocysteinemia; RNA-seq, RNA-sequencing; MC, monocyte; Cbs, Cystathionine β-synthase; SDE, significant differentially expressed; IPA, Ingenuity Pathway Analysis, TF, transcription factor, MΦ, macrophage.
