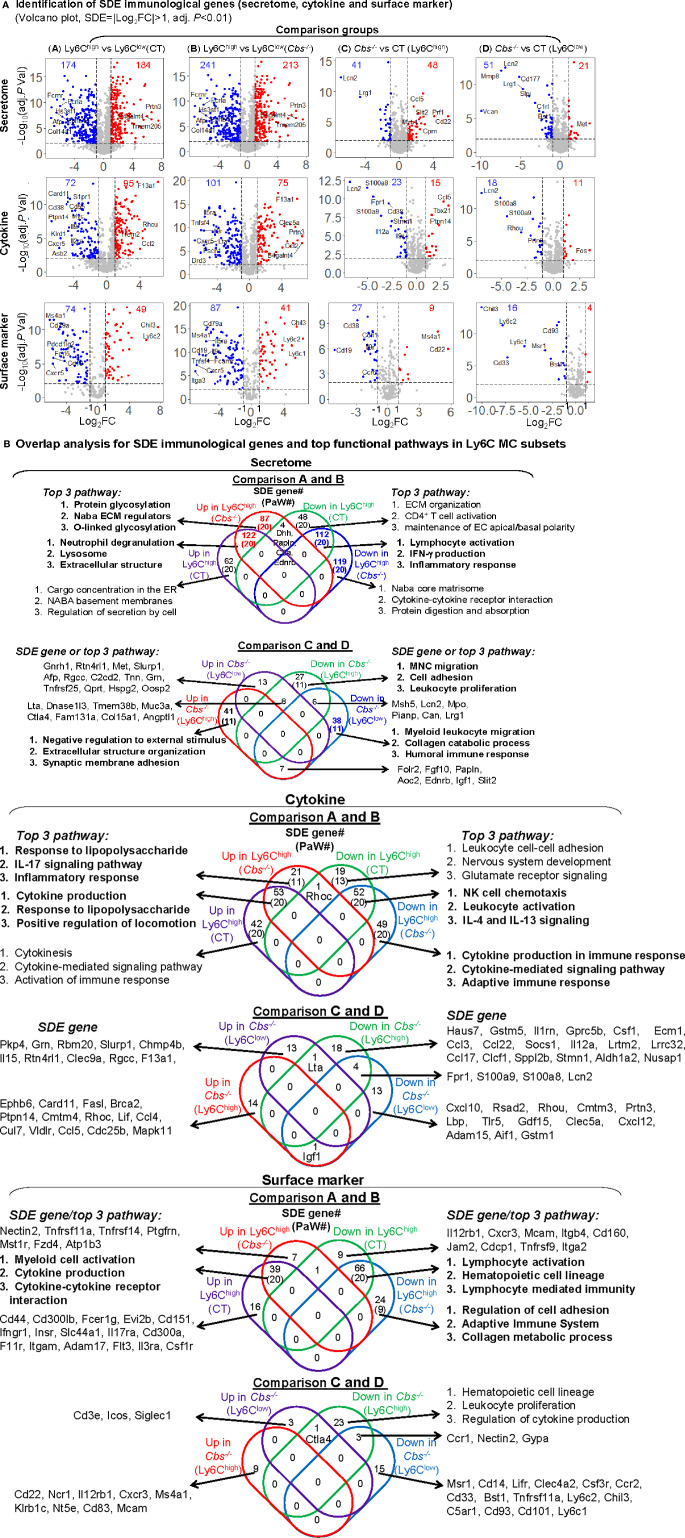
Figure 4.
Immunological signature genes and top functional pathways in Ly6C MC subset from CT and Cbs -/- mice. (A) Identification of immunological SDE genes (secretome, cytokine and surface marker). Volcano plot of all genes demonstrates the expression pattern of SDE genes in four comparison groups. Down-regulated SDE genes are highlighted in green and up-regulated in red (|Log2FC|>1, adj. P<0.01), with Log2FC as x-axis and −Log10(adjust P-value) as y-axis. SDE secretome, cytokine and surface marker were identified using the immunological gene set established in our previous study (PMID: 32179051) from website (https://www.proteinatlas.org/). Top 25 up- and down-regulated SDE genes in all comparisons via IPA are listed in Supplementary Table 2 . (B) Overlap analysis for SDE immunological genes in Ly6C MC subsets and top pathways. Venn diagram summarized the total SDE genes and their top three pathways in each SDE set from four pairs of comparisons. Functional pathways were developed by metascape software mainly using the GO database only in SDE set (>20 SDE genes). The top 3 functional pathways are presented. Numbers depict the amount of SDE genes. Numbers in the parentheses describes the number of pathways. A detailed list of SDE genes and pathway in each SDE set are presented in Supplementary Table 3 . ECM, extracellular matrix; EC, extracellular; IFNγ, interferon gamma; MNC, mononuclear cell; NK, natural killer.