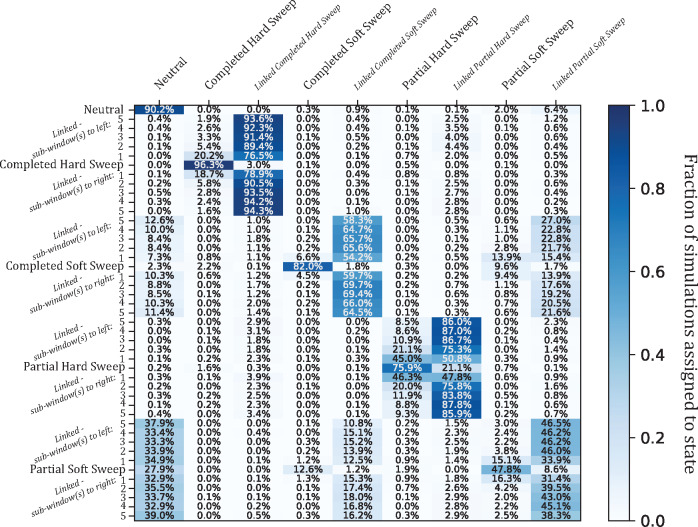
Fig. 2.
Confusion matrix heatmap of partialS/HIC simulation experiment. Given the BFS population history. Each row designates a true simulation class, which for linked sweeps is differentiated by distance in genomic subwindows to the target selective sweep. In total, there are 45 simulated scenarios shown, including 11 for each sweep type (i.e., one case whereby the sweep is within the central subwindow, and 10 whereby a linked sweep is located within one of the flanking subwindows) and neutrality. Each column indicates one of nine inferred states, with the linked simulation classes collapsed into a single category per selective sweep type for training and classification, allowing partialS/HIC to learn to distinguish between sweeps located in the focal subwindow and those somewhere nearby. Darker blue cells represent a higher proportion of the 1,000 calls for each true class. Importantly, there is generally high accuracy in discriminating between sweeps within the focal subwindow versus linked subwindows, especially when linkage is beyond one subwindow away and particularly for completed sweeps. However, discovering partial soft sweeps is noticeably a challenging task. Moreover, there is greater sensitivity separating full completion from partial completion for hard sweeps in contrast to soft sweeps.