Figure 1.
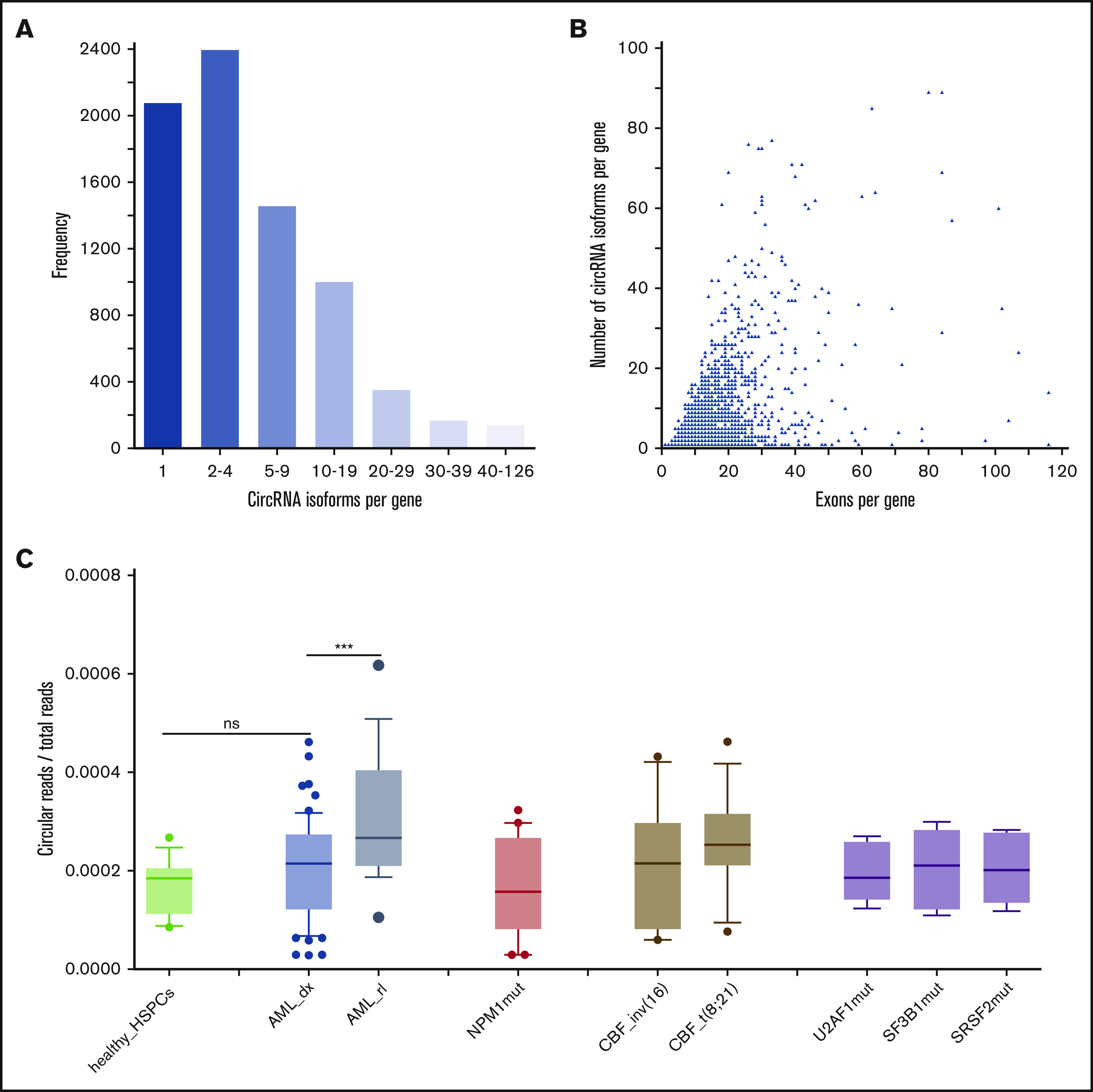
circRNA abundance in healthy HSPCs and AML. (A) Frequency of genes for which a certain amount of different circRNA isoforms could be detected across all diagnostic AML and control RNA-seq samples (n = 77). A total of 7578 circRNA-expressing genes was taken into account. (B) The number of different circRNA isoforms that were detected per gene in 77 RNA-seq samples in relation to the number of exons of the parental gene; 1924 highly expressed genes with a read count ≥ 20 were taken into account. (C) Ratio of total circular/linear read counts in RNA-seq data from 16 healthy HSPC samples (light green), 61 AML cases at diagnosis (blue), and 18 relapse cases (steel blue). Expression data were generated via ribosomal RNA–depleted RNA-seq, and reads were aligned and quantified using STAR. AML samples included diagnosis samples of patients with NPM1mut (red, n = 20); CBF leukemias (brown, n = 25), of which 11 carried inv(16) and 14 carried t(8;21); and 16 AML cases with mutations in splicing factors (lavender), including U2AF1 (n = 6), SF3B1 (n = 5), and SRSF2 (n = 6) (note that 1 case showed mutations in SF3B1 and SRSF2). The boxplots illustrate the median and interquartile range; whiskers denote the 10th to 90th percentiles. ***P < .001, unpaired Student t test. ns, not significant.
