Figure 1.
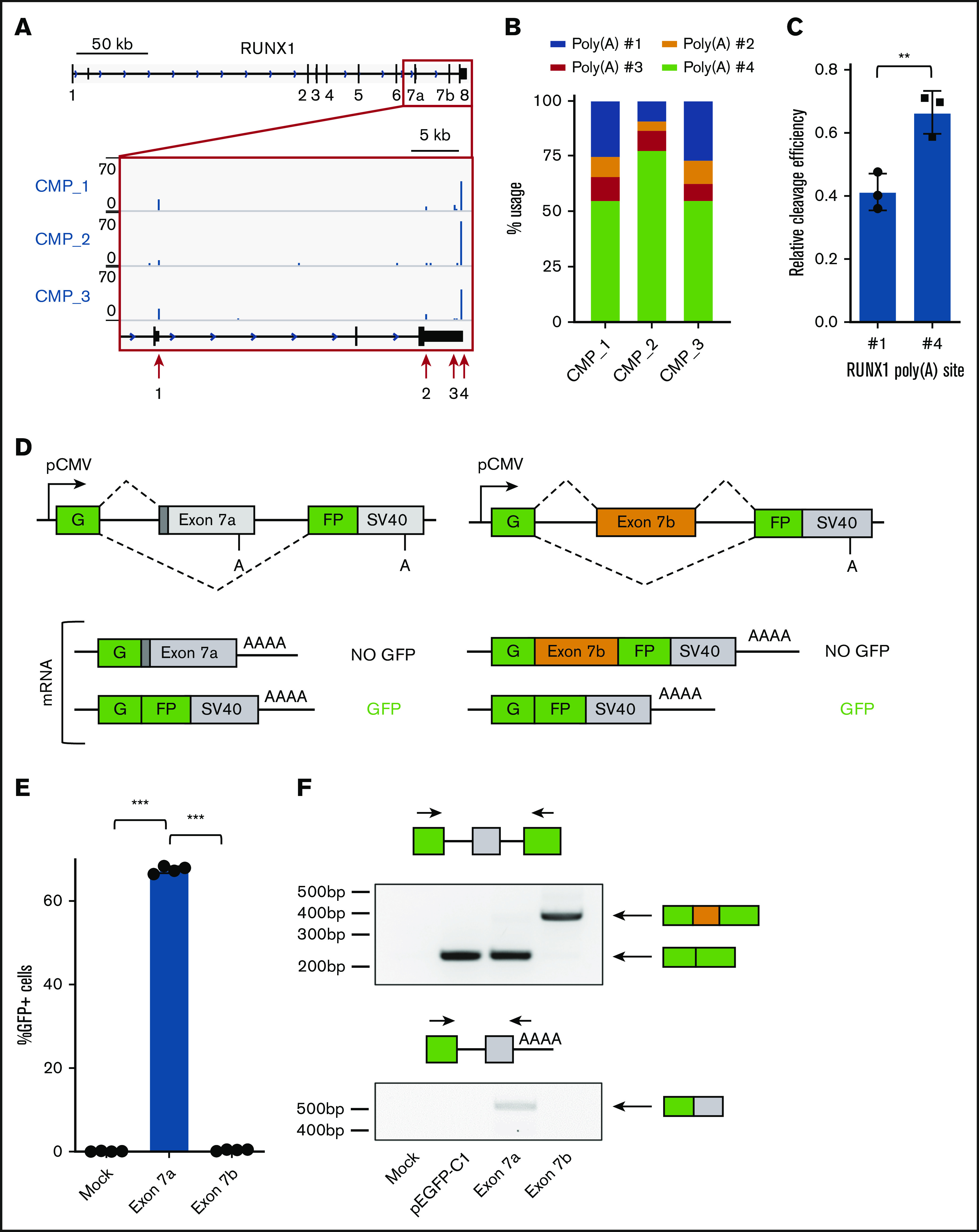
A split GFP minigene model recapitulates RUNX1 isoform generation. (A) Genome browser tracks depicting sequencing reads in the RUNX1 gene obtained from 3′READS of sorted common myeloid progenitors (CMP). The full RUNX1 genomic structure is shown above and the red boxed region is expanded below. The locations of the 4 major poly(A) sites are marked with red arrows. (B) Usage of the 4 major RUNX1 poly(A) sites, calculated from 3′READS analysis of sorted CMPs. (C) Quantification of RUNX1 poly(A) site #1 or #4 usage relative to a common synthetic poly(A) site via RNase protection assay. Data are mean ± standard deviation (SD) of 3 independent experiments. (D) Schematic diagram of the split GFP minigene constructs containing RUNX1 exon 7a (left panels) and RUNX1 exon 7b (right panels). Poly(A) sites are shown, as well as possible messenger RNA products. (E) Percentage of GFP+ cells, as measured by flow cytometry of KG-1a cells nucleofected with the RUNX1 exon 7a or exon 7b minigene construct. Data are mean ± SD of 4 independent experiments. (F) RT-PCR analysis of RNA extracted from KG-1a cells nucleofected with the RUNX1 minigene constructs, a GFP vector positive control, or mock-nucleofected negative control. Primer sets used for analysis are shown above the corresponding agarose gel image. **P < .01, 2-tailed Student t test; ***P < .001, 1-way analysis of variance (ANOVA) with a post hoc Tukey’s test.
