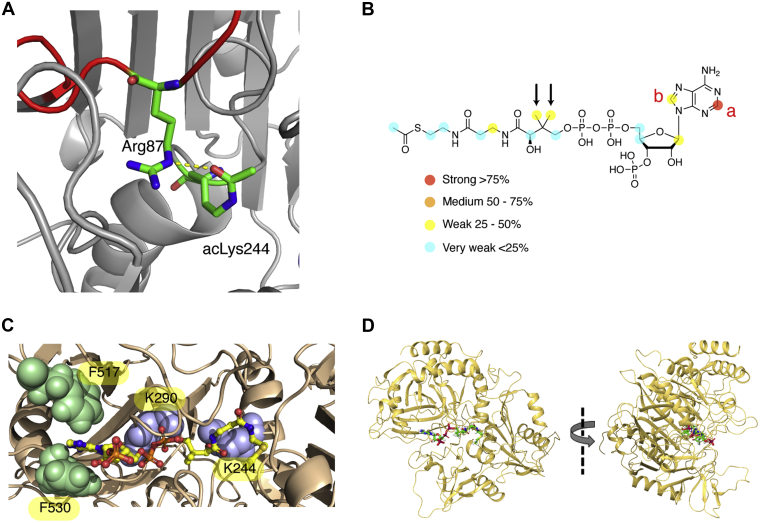
Figure 4.
Occupancy of acetyl-CoA inside the active site of PCK1 and determination of the effects of K244 acetylation.A, crystal structure of the PCK1 K244AcK mutant shows a hydrogen bond between acK244 and Arg87 that disrupts enzyme activity. The R-loop is highlighted in red. B, mapping of AcCoA-binding epitope upon interaction with PCK1 by STD NMR with saturation at −1.0 ppm. The colored spheres represent the normalized STD NMR intensity. Only STD responses are indicated for those protons that could accurately be measured. The letters a and b indicate the protons, and arrows, the methyl groups mentioned in the text. C, docking 3D model for the complex acetyl-CoA–PCK1. Detailed view of acetyl-CoA. The ligand is completely surrounded by PCK1 residues, in a narrow groove. Significantly, a π–πstacking involving uracyl is predicted with both F517 and F530, in agreement with STD-NMR results. D, full view of the docking complex, showing acetyl-CoA in the inner groove of the protein with the close side chains of K91 and K244. PCK1, phosphoenolpyruvate carboxykinase; STD, saturation-transfer difference.