Figure 6.
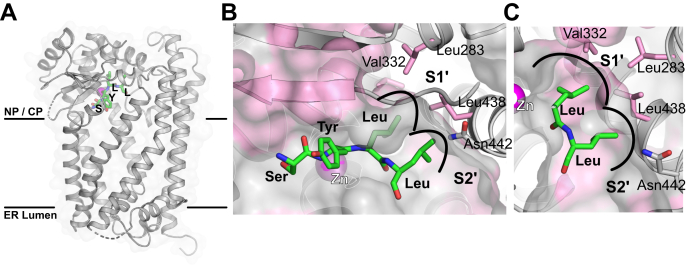
Model of the SYLL (P2–P2’ peptide) in the active site of ZMPSTE24.A, The overall structure of ZMPSTE24, determined by X-ray crystallography, showing position of the active site Zn2+ (magenta) and with the prelamin A –SYLL substrate positioned in the proposed catalytic site, based on the cocrystal structure of ZMPSTE24 and the CSIM tetrapeptide (31). Substrate peptide backbone carbons are labeled green, the nucleoplasmic-cytoplasmic (labeled NP/CP) and ER lumen sides of the membrane in which ZMPSTE24 sits are indicated. B, Zoom-in view of the active site, potential S1’ and S2’ binding pockets in the protease for P1’ and P2’ cleavage site residues of the substrate are annotated. C, View of the S1’ and S2’ binding sites showing the hydrophobic residues forming the prelamin A peptide binding sites. Hydrophobic residues are in pink and labeled with three-letter amino acid residue code.