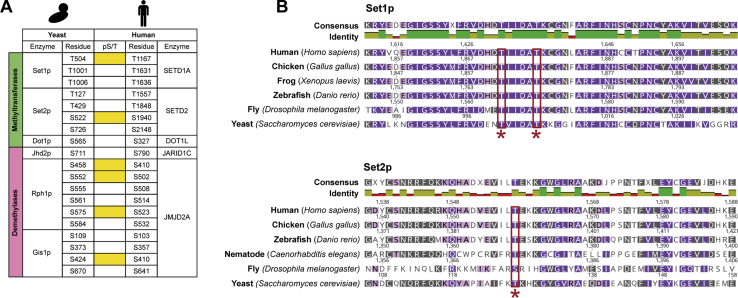
Figure 5.
Conservation of yeast phosphorylation sites on orthologous histone MTase and DMase enzymes.A, pairwise protein sequence alignments were performed between all yeast histone MTase and DMase enzymes and their respective human orthologs, with the exception of Set5p, which is not conserved in mammalian cells. Residues that are phosphorylated in S. cerevisiae and are conserved at the sequence level in human are tabulated. Conservation of phosphorylation at these residues is shown by yellow squares. B, representative portions of multiple sequence alignments for yeast Set1p (top) and Set2p (bottom) with several eukaryotic orthologs. Conservation of chemically similar amino acid residues is as follows: 100% = violet, 80 to 100% = mauve, 60 to 80% = khaki, < 60% = white. Residue numbers for each species are shown above their respective sequences. Instances where a phosphorylated serine or threonine residue on a yeast protein is conserved across all organisms are denoted by a red box and asterisk below the alignment. Sequence alignments were generated in Geneious Prime (version 2020.1.2).