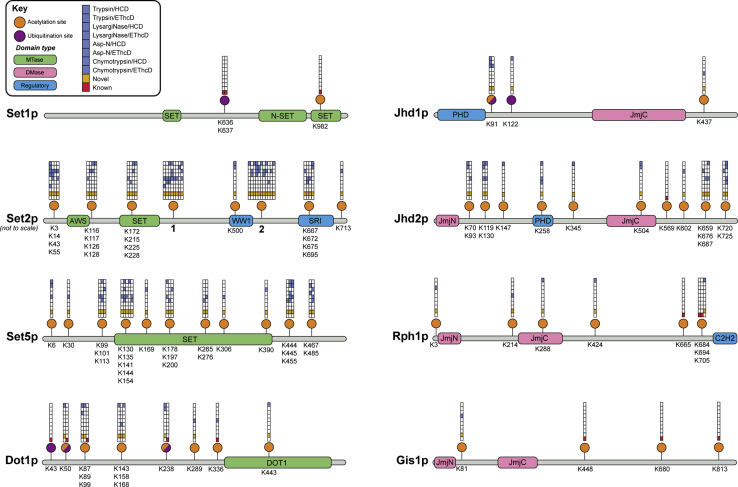
Figure 8.
Acetylation and ubiquitination sites on yeast histone MTases and DMases. Collation of acetylation and ubiquitination sites identified in this study and from previous high-throughput proteomic analyses (Tables S4 and S5). Acetylation (orange) and ubiquitination (purple) sites are displayed along linear protein sequence maps comprising MTase (green), DMase (pink), and other regulatory domains (blue). Sites that have evidence for both acetylation and ubiquitination are depicted as half orange/half purple circles. Grids above each PTM site give details about the site’s identification and are arranged according to the key (top left). Sites that have been previously reported in the literature are denoted by a red square, while novel sites identified in this study are shown with a gold square. Blue squares illustrate the identification of PTM sites using different protease/fragmentation method combinations. Where sites occur proximally to one another, grids are merged, and each column (left to right) corresponds to the modified residues listed below the protein map (top to bottom). To accommodate the large number of modification sites, Set2p is not illustrated to scale, and residue labels have been removed in instances where more than five grids have been merged (see numbering). These numbered grids correspond to the following residues from left to right: 1 = K329, K340, K412, K428, K433, K447, K450, K459; 2 = K510, K530, K541, K566, K574, K578, K584, K598, K602, K607, K620.