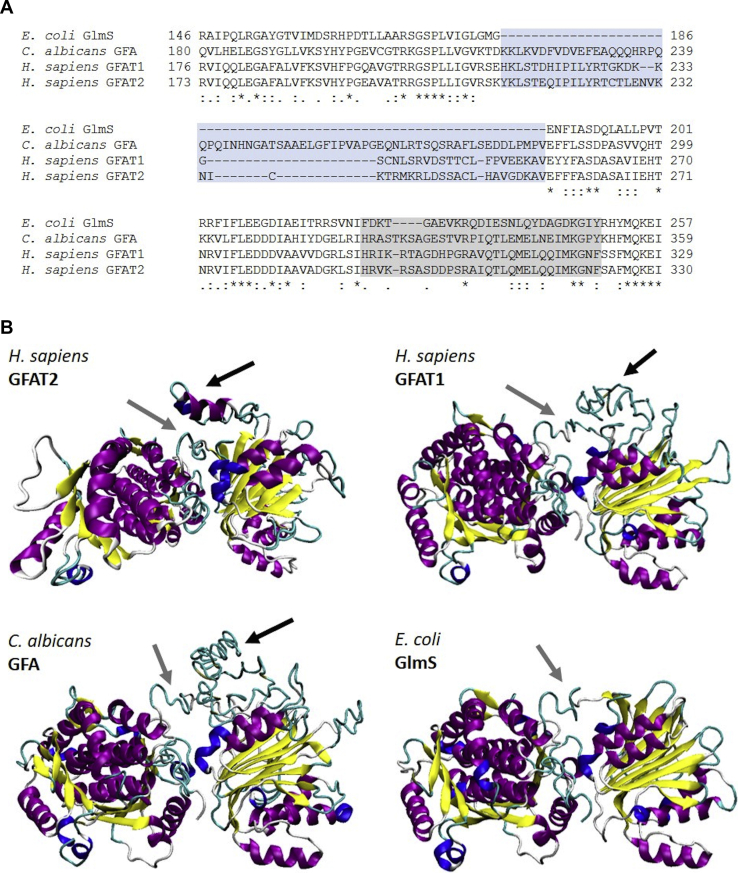
Figure 3.
Structure insights on human, fungal, and bacterial GFATs.A, alignment of the internal loop and interdomain connection sequences (highlighted in blue and gray, respectively) from GlmS (E. coli), GFA (C. albicans), and GFAT1 and GFAT2 (H. sapiens) performed with Clustal Omega server. Dashes indicate gaps, asteriscs indicate identical residues, and dots indicate residues with similar physical–chemical properties. B, three-dimensional models obtained for hGFAT2, hGFAT1, and GFA from C. albicans from threading using I-TASSER server. The structure of GlmS was retrieved from PDB under ID 4AMV. The proteins are represented in cartoon and colored according their secondary structure (α-helix in purple, β-sheet in yellow, 3–10 helix in blue, turns in cyan, and coil in white). The black arrows indicate the loop regions; the gray arrows indicate the interdomain region.