Figure 4.
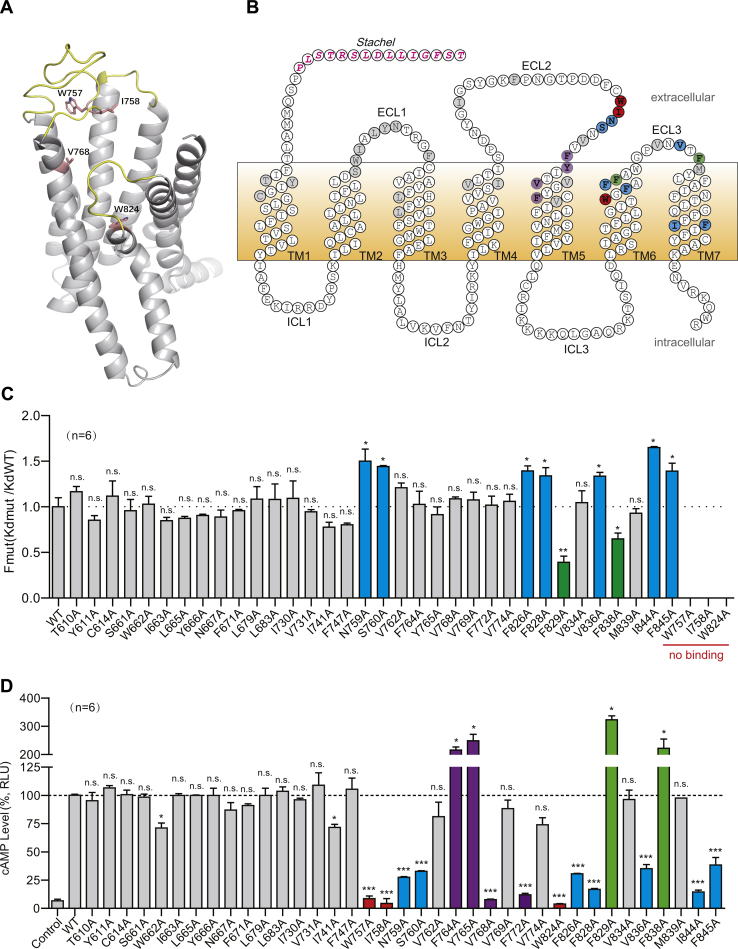
Key residues of ADGRG2 for VPM-p15 binding and signal transduction.A, a cartoon presentation of ADGRG2-ΔGPS-β highlighting the existence of the possible interactions between the VPM-p15 ligand and the binding site. The ADGRG2 structure was modeled by using PTH1R (Protein Data Bank: 6NBI) as a template. The extracellular loops are colored yellow and the ligand-binding residues are shown as side chain types and colored pink. B, a schematic serpentine representation of the ADGRG2 7TM domain residues highlighting its mutation sites. Extracellular and intracellular loops (ECL and ICL) are indicated (B). C, binding capacities of ADGRG2-ΔGPS-β WT or its mutants for VPM-p15 monitored by BRET experiments. HEK293 cells were transfected with Rluc-ADGRG2-ΔGPS-β or mutants. Transfected cells were stimulated by increasing the concentration of FITC-VPM-p15. Binding capacities were determined by BRET. Kd values of ADGRG2-ΔGPS-β WT and its mutants for binding VPM-p15 were calculated by GraphPad. n.s. p > 0.05, ∗p < 0.05, ∗∗p < 0.01, Binding capacities of ADGRG2-ΔGPS-β mutants were compared with ADGRG2-ΔGPS-β WT. D, effects of ADGRG2-ΔGPS-β mutants on VPM-p15-induced cAMP accumulation. HEK293 cells transfected with ADGRG2-ΔGPS-β or its mutants were stimulated by 100 μM VPM-p15. cAMP levels were detected by the Glosensor assay. Data were normalized by paralleling experiments with ADGRG2-ΔGPS-β WT. n.s. p > 0.05; ∗p < 0.05; ∗∗∗p < 0.001; ADGRG2-ΔGPS-β mutants were compared with ADGRG2-ΔGPS-β WT. C–D, each experiment was repeated six times.