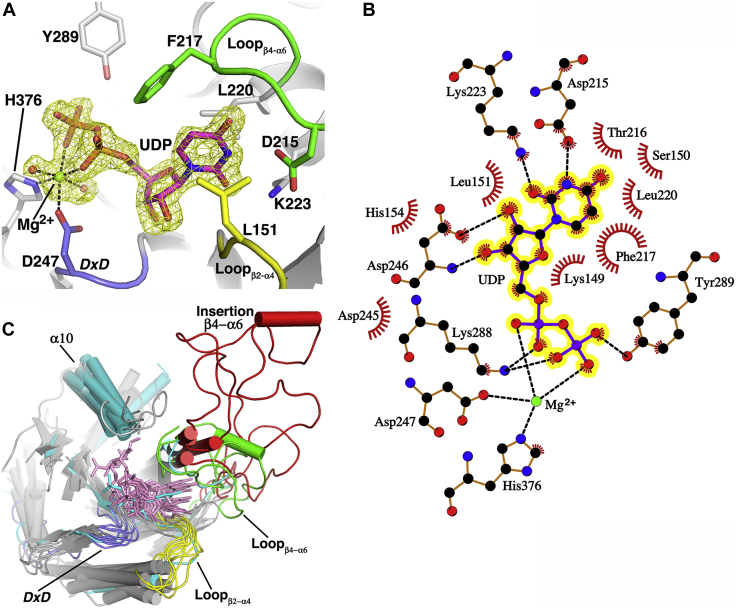
Figure 4.
A, Difference density map (Fo–Fc, yellow mesh) for the UDP and Mg2+ in the B3GNT2 nucleotide sugar binding pocket (pale gray cartoon) calculated at 1.55 Å and contoured at 3.5 σ. The map was calculated subsequent to the structure solution and an initial round of restrained refinement but prior to the modeling of the ligands. The UDP and Mg2+ are from the final refined coordinates of SeMet-B3GNT2:UDP:Mg2+ and colored as in Figure 3A. Amino acid side chains (stick mode) and structural elements Loopβ2-α4 (yellow), Loopβ4-α6 (green), and Loopβ5-β6 (DxD motif, slate blue) that interact with the UDP are shown. The octahedral coordination geometry of the metal ion involving the nucleotide diphosphate, side chains of H376 and D247 (last residue in the DxD motif), and solvent molecules (red spheres) is indicated with black dashes. B, Ligplot (79) representation of the B3GNT2 active site (orange, ball and stick) showing packing interactions (red, feathered lines) and hydrogen bonds (black, dashed lines) of the UDP (yellow highlight) and Mg2+ ion (green). C, The common active site architecture of GT-A fold glycosyltransferases (pale gray, cartoon) results in conformational similarity of bound nucleotide sugar donors and donor analogs (pink sticks). The donor binding pocket is shown with the structural elements Loopβ2-α4, Loopβ4-α6, and Loopβ5-β6 (DxD motif) that interact with the nucleotide colored as in Panel A. For some GT-A fold enzymes, Loopβ4-α6 can form extended regions (red) (e.g., GT6 (GGTA1), GT13 (MGAT1 and POMGNT1), and GT16 (MGAT2)). Helix α10, the location of the catalytic base in inverting GT-A fold GTs, is colored teal. The representative subset of GT-A fold structures from CAZy GT families 2, 6, 7, 13, 14, 16, 31, and 43 were aligned with B3GNT2 using the core GT-A fold (see Table S2).