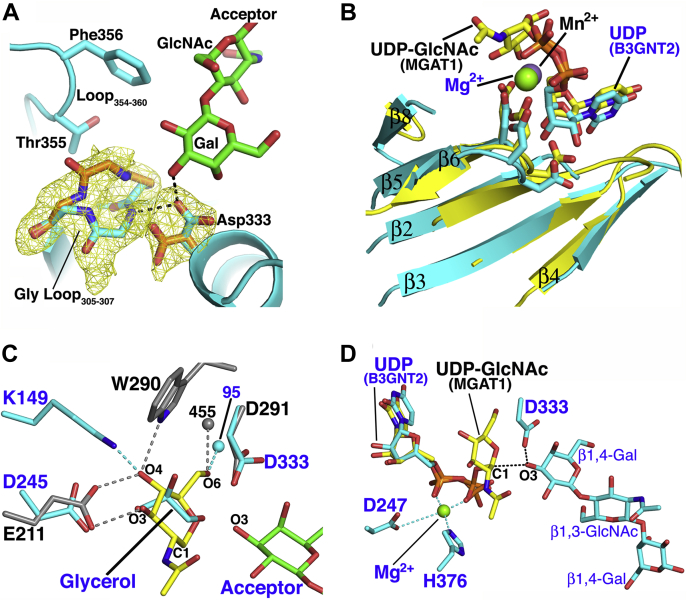
Figure 6.
Conformational changes in the B3GNT2 active site.A, Difference density map (Fo–Fc, yellow mesh) of the “Glycine-rich” loop (Gly-Loop305:307) and Asp333 in chain A of B3GNT2:UDP:Mg2+:LNnT (cyan cartoon), calculated at 1.85 Å and contoured at 3.0 σ. The map was calculated after omitting Gly-Loop305:307 and Asp333 from the refined coordinates of B3GNT2:UDP:Mg2+:LNnT and subjecting the model to simulated annealing. The active (cyan) and inactive (orange) conformations of Asp333 and Gly-Loop305:307 are shown. In the active conformation, the Asp333 carboxylate hydrogen bonds with the backbone amide of Gly303 and the nucleophile O3 of the acceptor Gal-β1,4- residue (black dashed lines). The nonreducing end Gal and GlcNAc of LNnT (green) and residue side chains from Loop354-360 that pack against the acceptor are also shown. B, the donor, UDP-GlcNAc (yellow, stick) modeled in the active site of B3GNT2:UDP:Mg2+:LNnT based on structural alignment with MGAT1:UDP-GlcNAc (1FOA). Superposition of the GT-A fold core β-sheets (β2, β3, β4, β5, β6, and β8) of MGAT1 (yellow, Mn2+ as purple sphere) and B3GNT2:UDP:Mg2+:LNnT (cyan, Mg2+ as green sphere) aligns the DxD motif (sticks), the metal ions, and the UDP moiety of the donor. C, the structural alignment of B3GNT2:UDP:Mg2+:LNnT (cyan) and MGAT1:UDP-GlcNAc (gray) to model the donor, UDP-GlcNAc, in the B3GNT2 active site. Hydrogen bonds between the MGAT1 side chains (sticks) and solvent water (gray sphere, numbered 455) and the GlcNAc (yellow, stick mode) are shown as gray dashed lines; the donor UDP has been omitted for clarity. In B3GNT2, a glycerol molecule occupies the same place as the modeled GlcNAc and the superposition places the O3 and O5 atoms of the GlcNAc in the same position as the O1 and O3 atoms of an ordered glycerol molecule. B3GNT2 residues (D245 and D333) and solvent water (cyan sphere, numbered 95) that are in position to conserve the hydrogen bonding interactions with the modeled GlcNAc are also shown. Interactions between O4 of the modeled GlcNAc and B3GNT2 residue K149 are depicted as dashed lines (cyan). D, The donor, UDP-GlcNAc (yellow, sticks), modeled in the active site of B3GNT2:UDP:Mg2+:LNnT (cyan). In B3GNT2, metal ion interactions involving the nucleotide diphosphate, side chains of H376 and D247 (last residue in the DxD motif) are depicted with cyan dashes. The proposed mechanism involves the catalytic base D333, deprotonating the O3 hydroxyl of the acceptor Gal-β1,4- residue. The deprotonation leads to the nucleophilic attack on the C1 atom of the UDP-GlcNAc donor (black dashed lines).