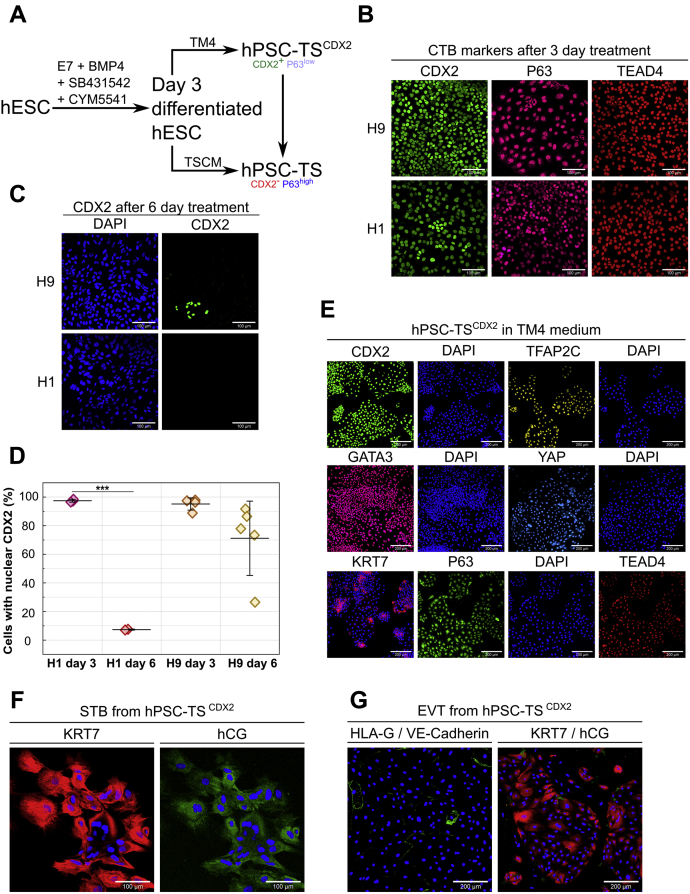
Figure 3.
Optimizing timing of hESC differentiation enables derivation of hPSC-TSCDX2cells. A, schematic of differentiation protocol for establishment of hPSC-TSCDX2 and hPSC-TS from hESCs. B, confocal images of 3 days treated H9 and H1 hESCs, staining for CDX2, P63, and TEAD4. Nuclei were stained with DAPI. The scale bars represent 100 μm. C, confocal images of 6 days treated H9 and H1 hESCs, staining for CDX2. Nuclei were stained with DAPI. The scale bars represent 100 μm. D, quantitative analysis of cells expressing nuclear CDX2 after 3- and 6-day differentiation treatment of H1 (day 3, 5455 cells in three images; day 6, 2448 cells in two images) and H9 (day 3, 5552 cells in four images; day 6, n = 6448 cells in five images) hESCs. Data points represent fraction of CDX2+cells in individual images from at least two biological replicates. Analysis was performed in MATLAB and at least two biological replicates were used. (Error bars are SD, ∗∗∗p < 0.05). E, confocal images of H9 hPSC-TSCDX2 in TM4, staining for CDX2, TFAP2C, GATA3, YAP, TEAD4, and P63. Nuclei were stained with DAPI. The scale bars represent 200 μm. F, confocal images of STB from H9 hPSC-TSCDX2 staining for hCG and KRT7. Nuclei were stained with DAPI. The scale bars represent 100 μm. G, confocal images of EVTs from H9 hPSC-TSCDX2, staining for HLA-G (red) and VE-Cadherin (green) as well as KRT7 (red) and hCG (green). Nuclei were stained with DAPI. The scale bars represent 200 μm. DAPI, 4′,6-diamidino-2-phenylindole; hESC, human embryonic stem cell; hPSC, human pluripotent stem cell; STB, syncytiotrophoblast; TS, trophoblast stem.