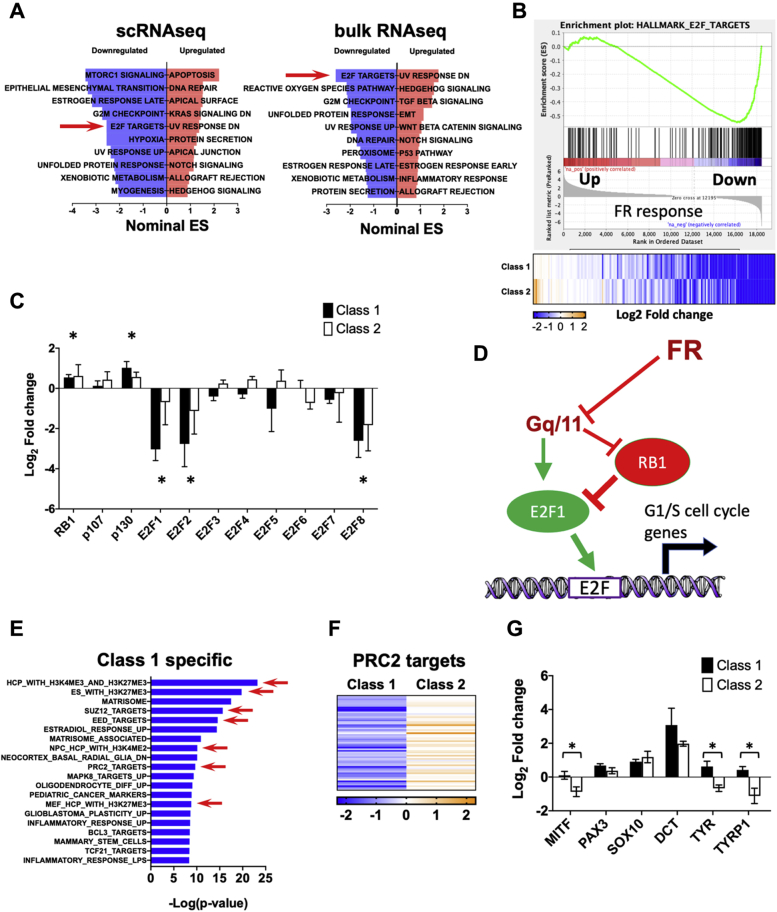
Figure 4.
Pathway analysis of FR-induced transcriptional responses in UM.A, hallmark gene-set enrichment analysis of differential gene expression in response to FR. Left panel shows gene sets affected by FR in enucleation-derived UM cells identified by scRNAseq. Right panel shows gene sets affected by FR in nine biopsy samples described in Figure 3 as detected by RNAseq. Red arrow indicates that E2F target genes were significantly downregulated in both scRNAseq and bulk RNAseq datasets. B, enrichment plot of the E2F target hallmark gene set detected by bulk RNAseq. Heatmap below shows insignificant difference in response of this gene set to FR in class 1 versus class 2 biopsy samples. C, effects of FR on expression of Rb/E2F family members in class 1 and class 2 biopsy samples detected by RNAseq (p107:RBL1; p130:RBL2). Asterisks indicate significant effect (p < 0.01; paired t-test) relative to vehicle controls. Differences between class 1 and class 2 samples were insignificant. D, proposed model for cell cycle regulation by constitutively active Gq/11 and FR. Signaling by constitutively active Gq/11 leads to repression of Rb expression and induction of E2F expression that drives UM cell proliferation. FR attenuates these effects to arrest cell cycle progression. E, differential responses of class 1 and class 2 tumor biopsy samples to FR. Gene-set enrichment analysis identifies genes downregulated preferentially in class 1 relative to class 2 tumor biopsy samples (log-rank t-test). Red arrows indicate PRC2-specific gene sets. F, heat map showing effects of FR on PRC2-targeted genes in class 1 versus class 2 tumor samples. G, responses of melanocytic differentiation genes to FR in class 1 and class 2 tumor biopsy samples. Asterisks indicate statistically significant (p < 0.01; paired t-test) effects of FR on the indicated melanocytic differentiation genes in class 1 and class 2 biopsy samples.