Figure 1.
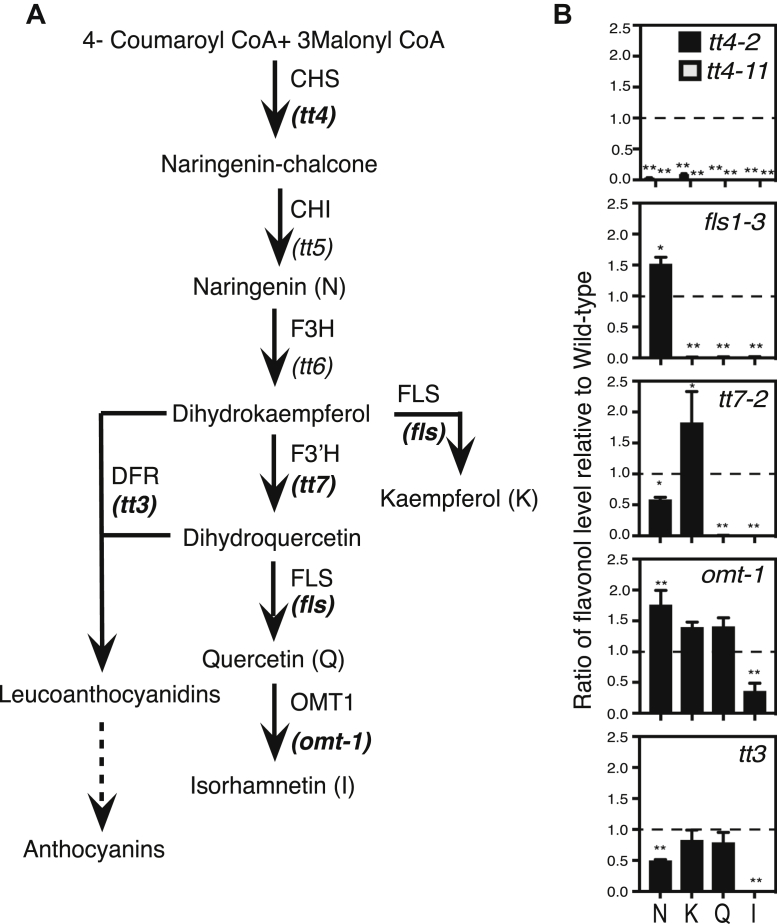
Flavonoid biosynthesis mutants have expected flavonol profiles.A, The flavonoid biosynthesis pathway in Arabidopsis has mutants at each step indicated in italics under the enzyme. Mutants in bold were used in the experiments in this manuscript. B, Flavonol levels were determined by LC-MS and the amount in nmole/gfw (gram fresh weight) was calculated. The ratio of the metabolite concentration in the mutant was divided by the wild-type levels reported in the mutants. A dashed line indicates a ratio of 1, which is when metabolites are equivalent to wild-type levels. Statistics were determined using a one-way ANOVA on the ratio values followed by a Dunnett's post-hoc test. Significance is indicated by ∗p < 0.05, ∗∗p < 0.005. Data was collected from three separate experiments with total n of Col-0 (n = 22), fls1-3 (n = 4), tt4-2 (n = 12), tt4-11 (n = 6), tt7-2 (n = 5), omt-1 (n = 7), and tt3 (n = 11).