Figure 4.
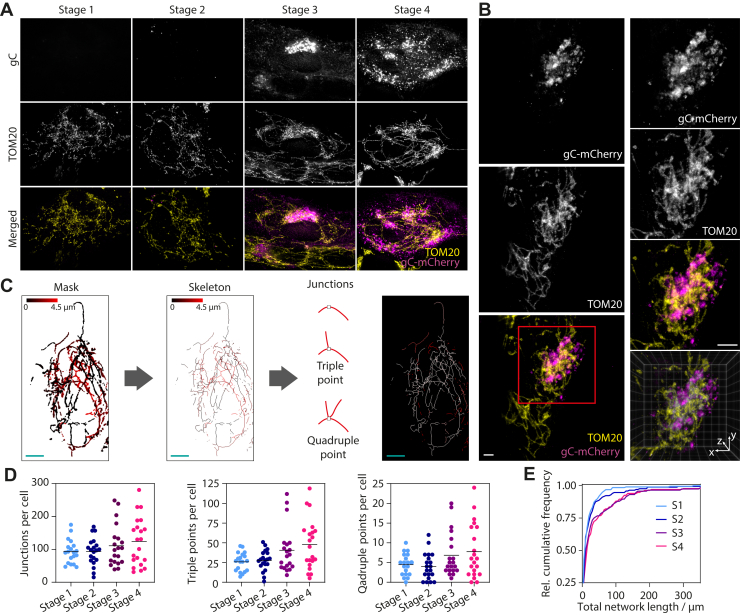
Interlacing of mitochondria and assembly compartments and spatial distribution of peroxisomes.A, HFF cells were infected with eYFP-ICP0/gC-mCherry HSV-1 and fixed at 3.5, 5.5, 7.5, and 9.5 hpi. Mitochondria were labeled by staining TOM20 following standard immunofluorescence protocol, mounted and z-stacks were taken at a widefield microscope. Displayed are maximum intensity projections after deconvolution showing exemplary cells for each stage of infection. From the top to bottom row: gC, mitochondria (TOM20) and the two channels merged (magenta: gC, yellow: mitochondria). B, infected cells were fixed 9 hpi and immunostained for TOM20 to label mitochondria. mCherry signal was enhanced by use of a nanobooster. Samples were then expanded using a published expansion microscopy protocol (42), and cells in stage 3 imaged using light sheet microscopy (Fig. S5). 3D structure of intertwined mitochondria and sites of secondary envelopment can be clearly observed. Upper row, whole cell, lower row, juxtanuclear region. C, for quantitative analysis of mitochondria networks, first masks were created from z-stacks as described in (A), which were then skeletonised. The lookup table (LUT) indicates the height of the mitochondria in z. The skeletons were analyzed to yield information about junctions and networks (LUT indicates individual mitochondria networks within the cell). D, scatter plots with indicated mean values show a slight increase in the number of junctions per cell for stages 3 and 4 compared with stages 1 and 2. A stronger increase is noticeable for triple points and quadruple points. E, as can be seen from plotting the relative cumulative frequency of the total network length, mitochondria networks increase in total length for stages 3 and 4 compared with stages 1 and 2. (Stage 1: n = 19 cells, stage 2: n = 20 cells, stage 3: n= 20 cells, stage 4: n = 21 cells for (D) and (E)). Scale bars 10 μm.