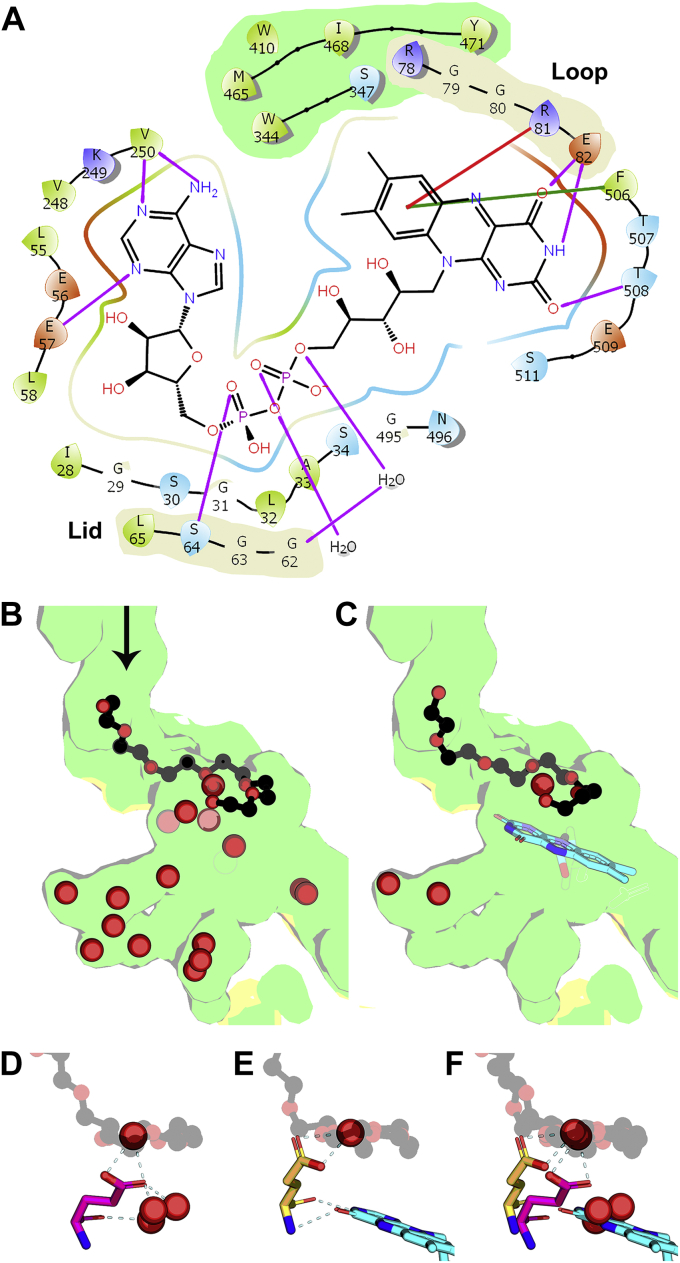
Figure 4.
Organization of the active site by FAD binding.A, Schematic of hydrogen bond (purple lines), π–π stacking (green line), and cation–π (red line) interactions between OhyA structural elements and FAD in the OhyA•PEG400•FAD complex with the lid closed (PDB: 7KAW, MolC) created with Maestro. The locations of the catalytic Loop and Lid are highlighted (beige shading). The hydrophobic surface accommodating the FAD xylene ring is derived from the fatty acid lobe (green shading). B, The active site before FAD binding in the OhyA•PEG400 complex (PDB: 7KAV, MolA). The active site is formed by the FAD (yellow) and fatty acid (green) lobes and contains the substrate analog PEG400 (black) and water molecules (red spheres). The arrow denotes direction of the incoming fatty acid. C, The active site with FAD bound in the OhyA•PEG400•FAD complex (PDB: 7KAW, MolB). Same orientation and color coding as Panel B but containing FAD (cyan). D, Glu82 (magenta) interacts with four ordered waters in the OhyA•PEG400 complex (PDB: 7KAV, MolA). E, Glu82 (yellow) coordinates a single water following FAD (cyan) binding in the OhyA•PEG400•FAD complex (PDB: 7KAW, MolB). Dashed lines are hydrogen bonds connecting FAD to the backbone carbonyl and amide of Glu82. F, Overlay of Panels D and E illustrating the desolvation of the active site, the conformational change in the orientation of Glu82, and the substrate water. FAD, flavin adenine dinucleotide; OhyA, oleate hydratase; PEG400, polyethylene glycol 400.