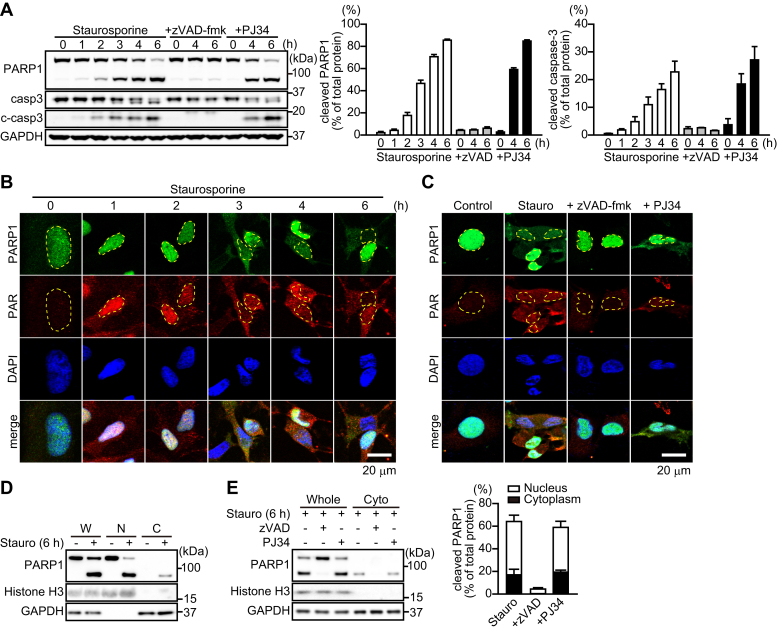
Figure 2.
PARP1 cleavage by caspase-3 results in the translocation of the 89-kDa PARP1 fragment to the cytoplasm.A, cleavage of PARP1 and caspase-3 (c-caspase-3) after staurosporine exposure. zVAD-fmk (50 μM) or PJ34 (10 μM) was added to the media for 30 min before 300 nM staurosporine exposure. Anti-PARP1 antibody recognizes full-length PARP1 and its 89-kDa PARP1 fragment. GAPDH was used as the loading control. The bar graphs represent cleaved PARP1 (left) and caspase-3 (right), respectively, assessed with pooled densitometric data (means ± S.E.M., n =3). Data are normalized to the ratio of cleaved proteins to total proteins. B, time-dependent change in the localization of PARP1 and PAR after 300 nM staurosporine exposure for indicated times. Cells were subjected to immunocytochemistry using anti-PARP1 antibody (green), anti-PAR antibody (red), and DAPI staining (blue). Yellow lines indicate the position of nuclei. The scale bar represents 20 μm. C, effect of caspase and PARP inhibition on PARP1 localization. zVAD-fmk (50 μM) or PJ34 (10 μM) was added for 30 min before 6-h exposure to 300 nM staurosporine. The scale bar represents 20 μm. D, subcellular localization of PARP1 after 6-h exposure to staurosporine (300 nM). Subcellular fractionation was performed to separate nuclei (N) and cytoplasmic (C) fractions. Histone H3 and GAPDH were used as nuclear and cytoplasmic markers, respectively. W indicates whole cells. E, effect of caspase and PARP inhibition on subcellular PARP1 localization. zVAD-fmk (50 μM) or PJ34 (10 μM) was added for 30 min before a 6-h exposure to staurosporine (300 nM). The bar graph represents the ratio of cleaved PARP1 to total PARP1 in nuclear (white) and cytoplasmic (black) fractions, assessed with pooled densitometric data (means ± S.E.M., n = 3). DAPI, 4',6-diamidino-2-phenylindole; PAR, poly(ADP-ribose); PARP1, poly(ADP-ribose) polymerase 1.