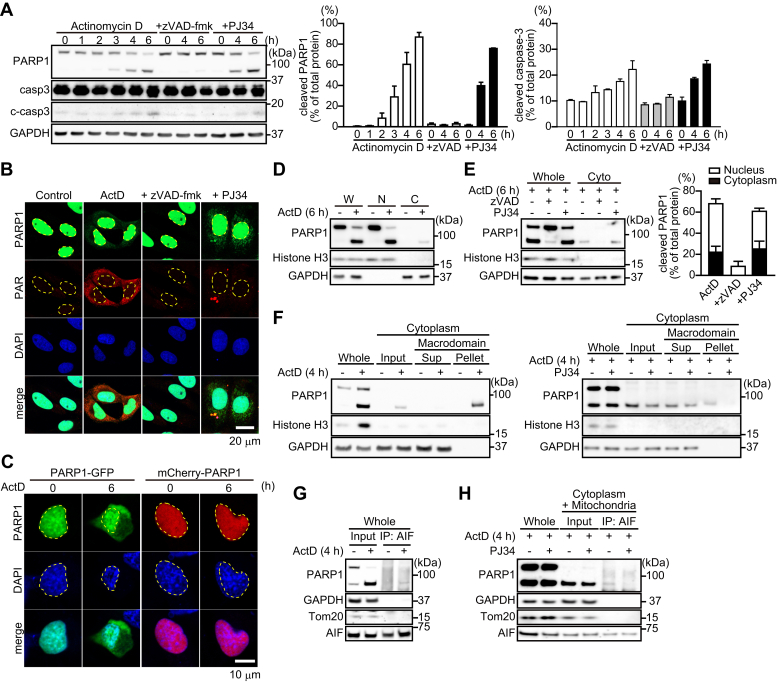
Figure 7.
Actinomycin D-induced PARP1 cleavage results in the translocation of 89-kDa PARP1 fragments to the cytoplasm and the binding to AIF via PAR polymers.A, PARP1 and caspase-3 cleavage after actinomycin D exposure. After exposure to actinomycin D (300 nM) for indicated times, cells were subjected to Western blotting with anti-PARP1 antibody and anticaspase-3 antibody. Anti-PARP1 antibody recognizes full-length PARP1 and 89-kDa PARP1 fragments. GAPDH was used as the loading control. The bar graphs represent cleaved PARP1 (left) and capase-3 (right), respectively, assessed with pooled densitometric data (means ± S.E.M., n = 3). Data are normalized to the ratio of cleaved proteins to total proteins. B, intracellular localization of PARP1 and PAR after 6-h exposure to actinomycin D (300 nM). Cells were subjected to immunocytochemistry using anti-PARP1 antibody (green), anti-PAR antibody (red), and DAPI staining (blue). Yellow lines indicate the position of nuclei. The scale bar represents 20 μm. C, localization of PARP1 constructs after 6-h exposure to actinomycin D. HeLa cells were exposed to actinomycin D (300 nM) for 6 h. Nuclei were stained with DAPI (blue). Yellow lines indicate the position of nuclei. The scale bar represents 10 μm. D, subcellular localization of PARP1 after 6-h exposure to actinomycin D (300 nM). Subcellular fractionation was performed to separate nuclei (N) and cytoplasmic (C) fractions. Histone H3 and GAPDH were used as nuclear and cytoplasmic markers, respectively. W indicates whole cells. E, effect of caspase and PARP inhibition on subcellular PARP1 localization. zVAD-fmk (50 μM) or PJ34 (10 μM) was added for 30 min before a 6-h exposure to actinomycin D (300 nM). The bar graph represents the ratio of cleaved PARP1 to total PARP1 in nuclear (white) and cytoplasmic (black) fractions, assessed with pooled densitometric data (means ± S.E.M., n = 3). F, identification of 89-kDa PARP1 fragments automodified by PAR in the cytoplasm. HeLa cells were exposed to actinomycin D (300 nM) for 4 h (left) without or with PJ34 (10 μM) (right). After subcellular fractionation, cytoplasmic fractions were subjected to GST pull-down assay using GST macrodomain. Histone H3 and GAPDH were used as nuclear and cytoplasmic markers, respectively. G, 89-kDa PARP1 fragments bind AIF. After 4-h exposure to actinomycin D (300 nM), cells were subjected to immunoprecipitation with anti-AIF antibody, followed by Western blotting using anti-PARP1 antibody. GAPDH was used for a loading control. H, 89-kDa PARP1 fragments bind AIF via PAR polymers in the cytoplasm. PJ34 (10 μM) was added for 30 min before 4-h exposure to 300 nM staurosporine. After subcellular fractionation, cytoplasmic/mitochondrial fractions were subjected to immunoprecipitation with anti-AIF antibody and then Western blotting using anti-PARP1 antibody. GAPDH and Tom20 were used as cytoplasmic and mitochondrial markers, respectively. DAPI, 4',6-diamidino-2-phenylindole; PARP1, poly(ADP-ribose) polymerase 1; PJ34, PARP inhibitor; zVAD, caspase inhibitor.