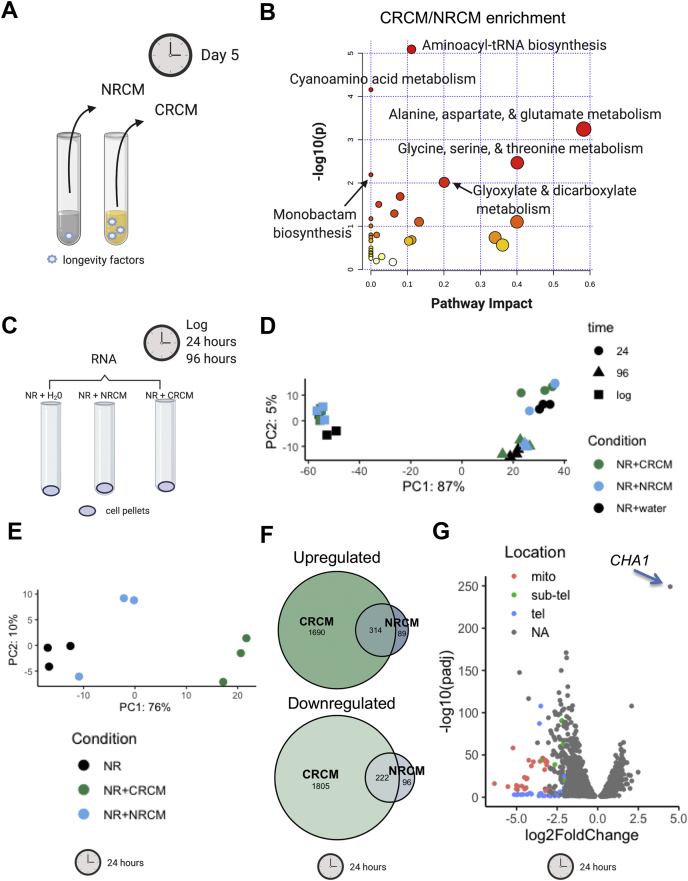
Figure 3.
Metabolomic and RNA-seq analysis point toward amino acid metabolism.A, schematic of conditioned medium collection used for untargeted metabolomics analysis after 5 days of SC cultures grown with 2.0% (NR) or 0.5% glucose (CR). B, metaboAnalyst software was used for Enrichment and Pathway Analysis of metabolites with a CR/NR ratio greater then 1.0. The names of KEGG pathways with p < 0.05 are highlighted. Pathway impact is a measure that considers the centrality of a metabolite in the pathway. Circle size is proportional to pathway impact value, and darker red color indicates more significant changes. C, schematic of cell conditions (NR, NR + 2.0 % NRCM, and NR + 2.0% CRCM) collected for RNA-seq analysis at 6 h (log phase), 24 h, and 96 h. Three biological replicates were performed for each condition. D, principal component analysis of RNA-seq samples at log, 24, and 96 h conditions of NR, NR+ NRCM, and NR+ CRCM. E, principal component analysis of RNA-seq samples collected at 24 h. F, Venn diagram of differentially expressed genes (up or down; FDR<0.05) for NRCM- or CRCM-supplemented samples, as compared with the NR + H2O control at 24 h. G, volcano plot displaying differential expressed genes between the NR + CRCM and NR + H2O samples at 24 h. The y-axis indicates the p-adjusted value, and x-axis the log2 fold change. Red, green, and blue denote genes located in the mitochondrial genome, subtelomeric, or telomeric regions, respectively. The most upregulated gene CHA1 is highlighted by an arrow.