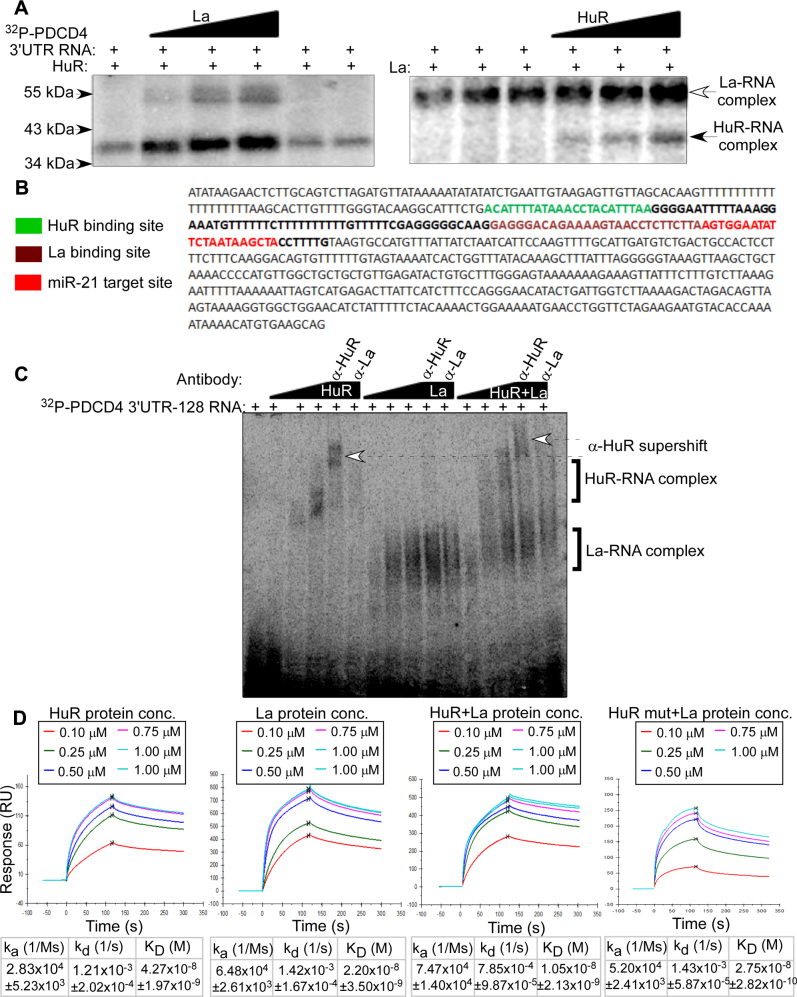
Figure 6.
La and HuR bind cooperatively to PDCD4 mRNA 3ʹUTR. A,32P-UTP-labeled PDCD4 3ʹUTR RNA was incubated with either a constant amount (20 pmole) of purified HuR protein in presence of increasing amounts (5, 10, and 20 pmole) of purified La protein (left panel) or with a constant amount of La protein (20 pmole) in presence of increasing amounts (5, 10, and 20 pmole) of HuR protein (right panel). The RNP complexes were UV-crosslinked, digested with RNase A, and resolved on 10% SDS-PAGE. Empty and filled arrows indicate the La and HuR RNP complexes, respectively. B, PDCD4 3ʹUTR sequence showing the 128 nt sequence (in bold) used for the REMSA containing the HuR binding site (green), La binding site (brown), and miR-21 binding site (red). C, RNA electrophoretic mobility shift assay (REMSA) using purified HuR and La proteins in three increasing concentrations (5, 10, and 20 pmole), either individually or in combination, with equal concentration of P32-labeled-PDCD4-3ʹUTR RNA 128 nt fragment shown in B. α-HuR and α-La lanes indicate reaction mixture having HuR and La antibody respectively for supershift assay. The shifted HuR-RNA and La-RNA complexes are indicated by second brackets, and complexes super-shifted with anti-HuR antibody are indicated by dotted arrows. D, 3ʹ-biotinylated PDCD4 3ʹUTR RNA was immobilized on the Biacore SA chip. Increasing concentrations of purified La and wildtype and mutant HuR proteins, either individually or together, were flown over the chip, and response units was plotted against time. The total amount of protein either flown individually, or when together, was same. The curves were fitted considering 1:1 binding kinetics, and binding constants (Ka, Kd, and KD) were calculated. The binding constants represent the mean ± SEM from three independent experiments. HuR, human antigen R; La, lupus antigen; PDCD4, programmed cell death 4; 3ʹUTR, 3ʹ-untranslated region.