Abstract
Lipid transfer proteins of the Ups1/PRELID1 family facilitate the transport of phospholipids across the intermembrane space of mitochondria in a lipid-specific manner. Heterodimeric complexes of yeast Ups1/Mdm35 or human PRELID1/TRIAP1 shuttle phosphatidic acid (PA) mainly synthesized in the endoplasmic reticulum (ER) to the inner membrane, where it is converted to cardiolipin (CL), the signature phospholipid of mitochondria. Loss of Ups1/PRELID1 proteins impairs the accumulation of CL and broadly affects mitochondrial structure and function. Unexpectedly and unlike yeast cells lacking the CL synthase Crd1, Ups1-deficient yeast cells exhibit glycolytic growth defects, pointing to functions of Ups1-mediated PA transfer beyond CL synthesis. Here, we show that the disturbed intramitochondrial transport of PA in ups1Δ cells leads to altered unfolded protein response (UPR) and mTORC1 signaling, independent of disturbances in CL synthesis. The impaired flux of PA into mitochondria is associated with the increased synthesis of phosphatidylcholine and a reduced phosphatidylethanolamine/phosphatidylcholine ratio in the ER of ups1Δ cells which suppresses the UPR. Moreover, we observed inhibition of target of rapamycin complex 1 (TORC1) signaling in these cells. Activation of either UPR by ER protein stress or of TORC1 signaling by disruption of its negative regulator, the Seh1-associated complex inhibiting TORC1 complex, increased cytosolic protein synthesis, and restored glycolytic growth of ups1Δ cells. These results demonstrate that PA influx into mitochondria is required to preserve ER membrane homeostasis and that its disturbance is associated with impaired glycolytic growth and cellular stress signaling.
Keywords: mitochondria, endoplasmic reticulum, phospholipid, lipid transfer, Ups1, PRELID1, unfolded protein response, TORC1, yeast
Abbreviations: CL, cardiolipin; ER, endoplasmic reticulum; IM, inner membrane; OM, outer membrane; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; qMS, quantitative mass spectrometry; TORC1, target of rapamycin complex 1; UPR, unfolded protein response; UPRE, UPR element
The functional integrity of mitochondria depends on characteristic lipid compositions of their membranes, the mitochondrial outer and inner membrane (OM and IM) (1, 2). This is exemplified by the hallmark lipid of mitochondria, cardiolipin (CL), which is pivotal for the structure and function of mitochondria. CL maintains respiration and cristae morphogenesis, ensures protein biogenesis and affects the fusion and fission of mitochondrial membranes (3, 4, 5, 6). Altered CL levels in the OM modulate mitophagy and apoptosis (7, 8, 9, 10, 11). Hence, reduced levels and aberrant acylation or peroxidation of CL compromise mitochondrial activities and are associated with aging and various pathophysiological conditions, including cardiomyopathies, skeletal myopathies, ataxias, or nonalcoholic fatty liver disease (12, 13, 14, 15).
CL is synthesized along an enzymatic cascade at the IM from phosphatidic acid (PA), which is mainly synthesized in the endoplasmic reticulum (ER) and imported into mitochondria (16). Although it has been reported that PA can be produced in mitochondrial membranes by phospholipases and kinases (17, 18), the maintenance of mitochondrial lipid homeostasis requires extensive exchange of phospholipids between the ER and mitochondria (19, 20). Similar to other membrane lipids, PA is transported from the ER to the OM at membrane contact sites, which facilitate the bi-directional transport of phospholipids between both organelles. Studies on the cellular distribution of newly synthesized phosphatidylserine (PS) revealed that phospholipid transfer to mitochondria can be driven by localized synthesis (21, 22). After reaching the OM, phospholipids are transported across the intermembrane space by conserved, lipid-specific lipid transfer proteins (23, 24, 25, 26, 27, 28), which likely act in concert with MICOS membrane tethering complexes (24). Heterodimeric complexes of yeast Ups1 (human PRELID1) and Mdm35 (human TRIAP1) serve as PA-specific lipid transfer proteins (23, 26). Crystal structures of various members of the conserved Ups1/PRELID1 family of lipid transfer proteins show an internal lipid binding pocket, which is surrounded by a β-sheet and α-helices, reminiscent of other classes of lipid transfer proteins (29, 30, 31, 32). Loss of Ups1 or PRELID1 in yeast or human cells, respectively, inhibits intramitochondrial PA transport and limits CL synthesis at the IM, which is accompanied by mitochondrial deficiencies, such as impaired respiration and mitochondrial fragmentation, and increases the susceptibility of the cells toward apoptotic stimuli (23, 26, 33, 34, 35).
High levels of PRELID1 and TRIAP1 were identified as an unfavorable prognostic marker in cancer (36). As cancer cells are often glycolytic, it is noteworthy that yeast cells lacking Ups1 also exhibit slow growth in glucose media (23, 34, 35). This phenotype is difficult to reconcile with reduced CL levels only, as cells lacking the CL synthase Crd1 in mitochondria grow normally in glucose-containing media although they are virtually devoid of CL (37). It therefore appears that the inhibition of intramitochondrial PA transport upon loss of Ups1 does not only perturb the mitochondrial membrane homeostasis but also causes deficiencies presumably beyond mitochondrial functions. Here, we demonstrate that a disturbed intramitochondrial transport of PA affects TORC1 signaling and alters the phospholipid homeostasis of the ER membrane modulating the unfolded protein response (UPR).
Results
The loss of Ups1 impairs basal UPR
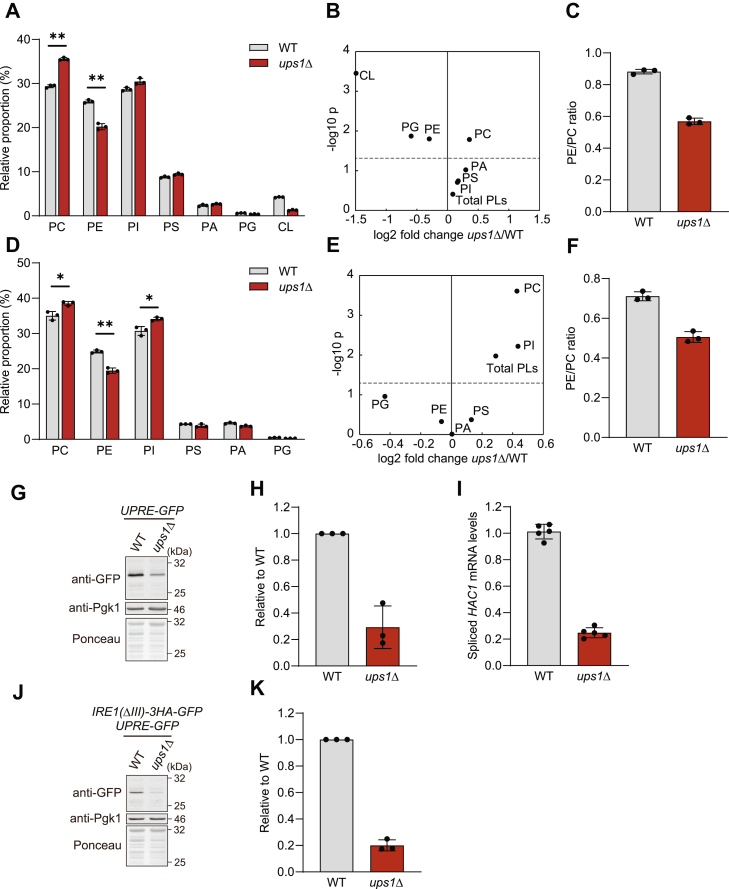
To examine how the loss of Ups1 affects cell growth under glycolytic conditions, we used quantitative mass spectrometry (qMS) to determine the phospholipid profile of cellular membranes in wild-type and ups1Δ cells which were grown in glucose-containing media. In agreement with previous findings (23, 34), we observed reduced CL levels in ups1Δ cells, reflecting impaired PA transport to CL-synthesizing enzymes at the IM (Fig. 1, A and B and Fig. S1A). We also noted a significant increase of phosphatidylcholine (PC) and a decrease of phosphatidylethanolamine (PE), resulting in a low PE/PC ratio in cellular membranes (Fig. 1, A–C and Fig. S1A). Unexpectedly, deletion of UPS1 also affected the phospholipid composition of ER membranes. We observed an increased total phospholipid content of an ER-enriched membrane fraction, which was accompanied by a significant increase of PC and phosphatidylinositol (PI) (Fig. 1, D and E and Fig. S1B), and a low PE/PC ratio (Fig. 1F).
Figure 1.
ER phospholipid composition is altered in ups1Δ cells. A, phospholipid composition in whole cell of wild-type (WT) and ups1Δ cells grown to log phase in SCD medium. Data represent mean ± SD (n = 3). ∗∗p < 0.01. B, changes in the absolute abundance of phospholipids in whole cell of ups1Δ. Data represented as log2 fold change at x-axis with −log 10 p value of student's t test at y-axis. Dashed line represents p = 0.05. C, PE/PC ratio of WT and ups1Δ cells. D, phospholipid composition in ER-enriched microsome fraction from WT and ups1Δ cells grown to log phase in SCD medium. Data represent mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01. E, changes in the absolute abundance of phospholipids in ER-enriched microsome fraction of ups1Δ. Data represented as log2 fold change at x-axis with −log 10 p value of Student's t test at y-axis. Dashed line represents p = 0.05. F, PE/PC ratio in ER-enriched microsome fraction from WT and ups1Δ cells. G, WT and ups1Δ cells expressing 4xUPRE-GFP grown to log phase in SCD medium were subjected to Western blotting. Pgk1 and ponceau staining were monitored as a loading control. H, GFP in (G) was quantified. GFP signals were normalized to Pgk1 and expressed relative to WT cells (set as one). Data represent mean ± SD (n = 3). I, WT and ups1Δ cells were grown to log phase in SCD medium. Spliced HAC1 mRNA expression was analyzed by real time PCR and normalized to ACT1 mRNA expression. Data represent mean ± SD (n = 5). J, WT and ups1Δ cells expressing Ire1(ΔIII)-3HA-GFP and 4xUPRE-GFPgrown to log phase in SCD medium were subjected to Western blotting. K, GFP in (J) was quantified as described in (H). Data represent mean ± SD (n = 3). ER, endoplasmic reticulum; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SCD, synthetic complete glucose; UPRE, UPR element.
Previous studies have reported that an increased PE/PC ratio in ER membranes activates the UPR independent of protein stress (38, 39, 40, 41). We therefore monitored UPR induction in ups1Δ cells harboring GFP under the control of the UPR element (UPRE) in their genome (42). Whereas GFP accumulated in wild-type cells under basal conditions, GFP expression was decreased in cells lacking Ups1 which were grown in glucose- (Fig. 1, G and H) or galactose-containing media (Fig. S1, C and D). Moreover, we observed reduced mRNA levels of spliced HAC1, encoding a transcription factor involved in UPR (Fig. 1I) (43, 44, 45). We therefore conclude that basal gene expression under the control of UPRE is suppressed in ups1Δ cells. Notably and in contrast to ups1Δ cells, cells lacking other enzymes involved in CL synthesis (such as Tam41, Pgs1, or Crd1) did not show significant reduction of the GFP expression under UPRE (Fig. S1, E–H), pointing that CL defect per se does not lead to the suppression of UPR.
A central sensor of UPR is the ER kinase Ire1, which senses unfolded proteins in the ER lumen as well as lipid bilayer stress in the membrane (43, 46, 47, 48). To assess the contribution of lipid bilayer stress specifically, we exploited the previously described ΔIII mutant of Ire1, which is able to sense lipid bilayer stress but not unfolded ER proteins (49, 50). Cells expressing Ire1(ΔIII) recapitulated the suppression of the UPR by the loss of Ups1 (Fig. 1, J and K). These results suggest that the altered phospholipid composition of ER membranes limits UPR activation in ups1Δ cells.
Increased PC levels cause suppression of basal UPR
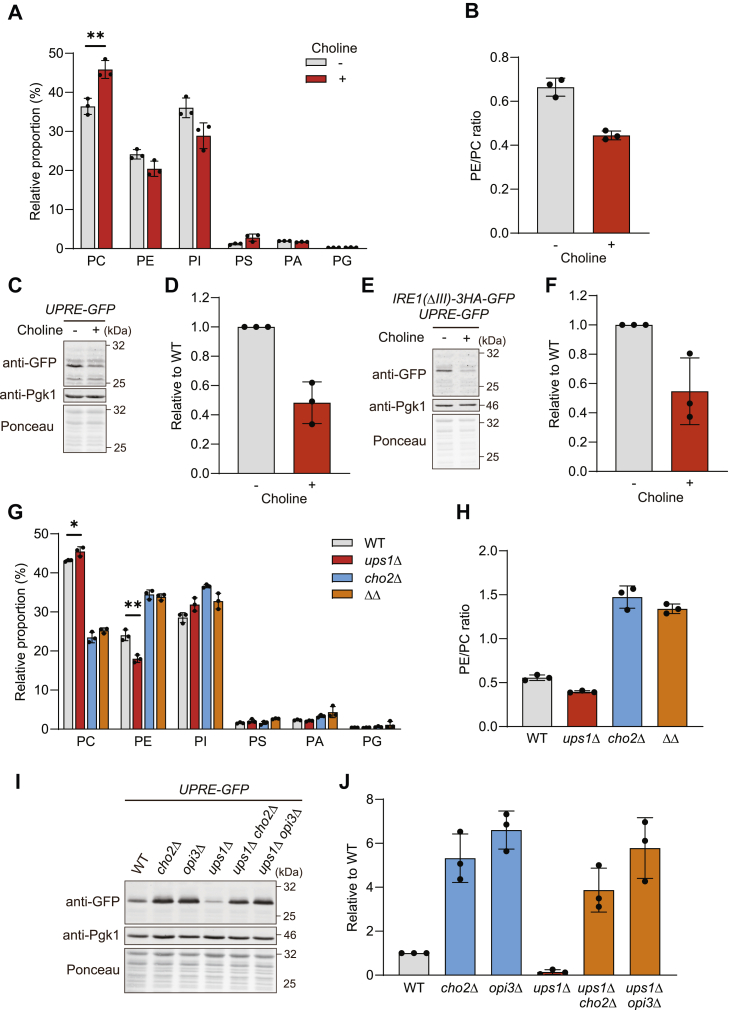
To examine how a decreased PE/PC ratio in ER membranes affects UPR, we induced de novo synthesis of PC via the Kennedy pathway by supplementing the growth medium with choline (51) and monitored UPR activity. Choline supplementation increased PC levels in ER-enriched membrane fractions of wild-type cells and led to a lower PE/PC ratio in these membranes (Fig. 2, A and B). Similar to cells lacking Ups1, the decreased PE/PC ratio in ER membranes was accompanied by an impaired basal UPR (Fig. 2, C and D), which was not altered in cells expressing mutant Ire1(ΔIII) (Fig. 2, E and F). We conclude from these experiments that the basal UPR is impaired if the PE/PC ratio in ER membranes is decreased, whereas an increased PE/PC ratio in ER membranes induces the UPR (41).
Figure 2.
Basal UPR is suppressed in high PC levels. A, phospholipid composition in ER-enriched microsome fraction from wild-type (WT) cells grown to log phase in SCD medium with or without 1 mM choline. Data represent mean ± SD (n = 3). B, PE/PC ratio in ER-enriched microsome fraction from WT with or without choline. C, WT cells expressing 4xUPRE-GFP grown to log phase in SCD medium with or without 1 mM choline were subjected to Western blotting. Pgk1 and ponceau staining were monitored as a loading control. D, GFP in (C) was quantified. GFP signals were normalized to Pgk1 and expressed relative to WT cells (set as one). Data represent mean ± SD (n = 3). E, WT cells expressing Ire1(ΔIII)-3HA-GFP and 4xUPRE-GFP grown to log phase in SCD medium with or without 1 mM choline were subjected to Western blotting. F, GFP in (E) was quantified as described in (D). Data represent mean ± SD (n = 3). G, phospholipid composition in ER-enriched microsome fraction from WT, ups1Δ, cho2Δ and ups1Δcho2Δ (ΔΔ) cells grown to log phase in SCD medium. Data represent mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01. H, PE/PC ratio of ER-enriched microsome fraction from WT, ups1Δ, cho2Δ and ups1Δcho2Δ (ΔΔ) cells. I, WT, cho2Δ, opi3Δ, ups1Δ, ups1Δcho2Δ, and ups1Δopi3Δ cells expressing 4xUPRE-GFP grown to log phase in SCD medium were subjected to Western blotting. J, GFP in (I) was quantified as described in (D). Data represent mean ± SD (n = 3). ER, endoplasmic reticulum; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SCD, synthetic complete glucose; UPR, unfolded protein response; UPRE, UPR element.
To unambiguously demonstrate that Ups1 affects UPR activation by modulating the phospholipid profile in ER membranes, we manipulated the PE/PC ratio in ups1Δ cells genetically. Yeast cells lacking Cho2 or Opi3, which are required for synthesis of PC from PE (51), accumulate PE relative to PC (Fig. 2G). Accordingly, deletion of CHO2 or OPI3 induces the UPR (41). We therefore deleted CHO2 or OPI3 in ups1Δ cells, which strongly increased the PE/PC ratio in an ER-enriched membrane fraction (Fig. 2, G and H). This was accompanied by UPR activation in the absence of Ups1 (Fig. 2, I and J). Thus, ups1Δ cells maintain the ability to induce the UPR upon lipid bilayer stress, but the reduced PE/PC ratio in the ER membrane of ups1Δ cells impairs the UPR under basal conditions.
UPR activation restores glycolytic growth of ups1Δ cells
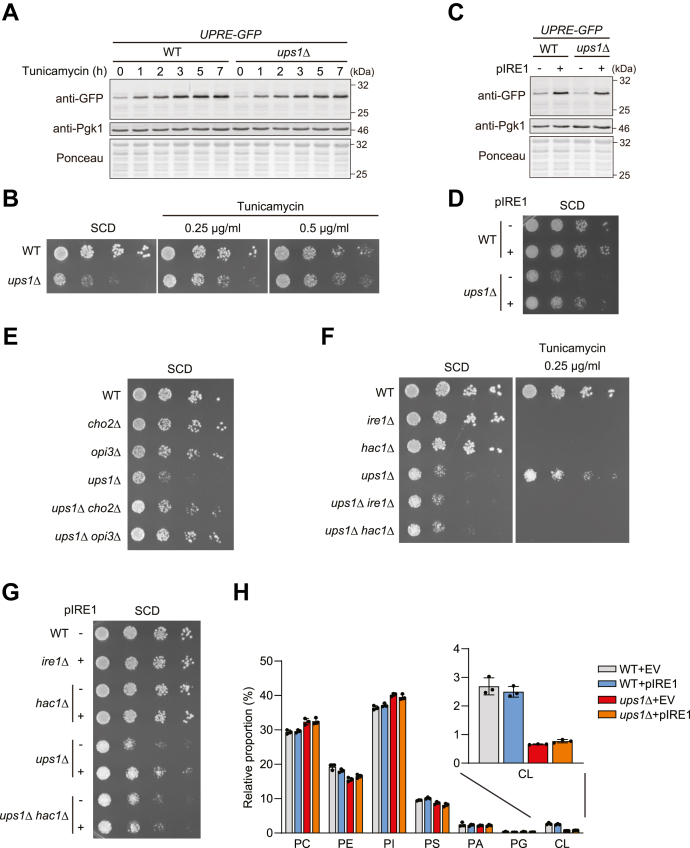
Having established the link between Ups1 and UPR activation, we examined in further experiments whether the reduced UPR activity can explain the impaired glycolytic growth of ups1Δ cells. We therefore treated ups1Δ cells with tunicamycin, an inhibitor of protein glycosylation, which causes ER protein stress and induces the UPR. Treatment with tunicamycin activated the UPR both in wild-type and in ups1Δ cells (Fig. 3A) and allowed growth of ups1Δ cells on fermentable medium (Fig. 3B). Consistently, activation of the UPR upon overexpression of Ire1 (Fig. 3C) (47, 52) or deletion of CHO2 or OPI3 (Fig. 2, I and J) suppressed growth deficiencies of ups1Δ cells on glucose-containing medium (Fig. 3, D and E). The restoration of cell growth by tunicamycin and overexpression of Ire1 depended on Ire1-Hac1 signaling (Fig. 3, F and G), demonstrating that UPR activation is sufficient to promote glycolytic growth of ups1Δ cells. Notably, previous studies correlated the growth of ups1Δ cells on glucose-containing media with mitochondrial CL levels (53). However, overexpression of Ire1 did not alter the accumulation of CL (Fig. 3G). We therefore conclude that UPR activation is sufficient to allow growth of ups1Δ cells on fermentable carbon sources independent of CL levels in mitochondrial membranes.
Figure 3.
Glycolytic growth of ups1Δ cells is restored upon UPR activation. A, wild-type (WT) and ups1Δ cells expressing 4xUPRE-GFP grown to log phase in SCD medium were treated with 1 μg/ml tunicamycin, collected at the indicated time points, and subjected to Western blotting. Pgk1 and ponceau staining were monitored as a loading control. B, serial dilutions of WT and ups1Δ cells were spotted on SCD medium with or without tunicamycin (concentration as 0.25 or 0.5 μg/ml) and incubated at 30 °C for 2 days (n = at least 3). C, 4xUPRE-GFP expressing WT and ups1Δ cells transformed with an empty vector or a plasmid encoding Ire1 (pIRE1) were grown to log phase in SCD medium and subjected to Western blotting. D, serial dilutions of WT and ups1Δ cells transformed with an empty vector or pIRE1 were spotted on SCD medium and incubated at 30 °C for 2 days (n = at least 3). E, serial dilutions of WT, cho2Δ, opi3Δ, ups1Δ, ups1Δcho2Δ, and ups1Δopi3Δ cells were spotted on SCD medium and incubated at 30 °C for 2 days (n = at least 3). F, serial dilutions of WT, ire1Δ, hac1Δ, ups1Δ, ups1Δire1Δ, ups1Δhac1Δ cells were spotted on SCD medium with or without 0.25 μg/ml tunicamycin and incubated at 30 °C for 2 days (n = at least 3). G, serial dilutions of WT, ire1Δ, hac1Δ, ups1Δ, ups1Δhac1Δ cells transformed with an empty vector or pIRE1 were spotted on SCD medium and incubated at 30 °C for 2 days (n = 3). H, phospholipid composition in whole cell of WT and ups1Δ cells transformed with an empty vector or pIRE1. Data represent mean ± SD (n = 3). SCD, synthetic complete glucose; UPR, unfolded protein response; UPRE, UPR element.
Reduced cellular protein synthesis in ups1Δ cells
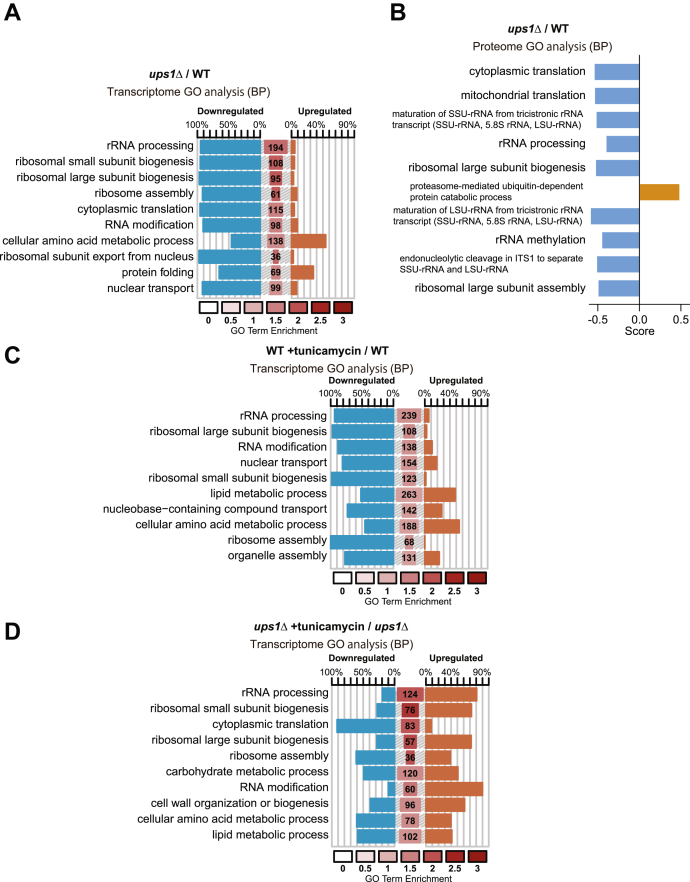
We noted that the loss of UPR activity in ire1Δ and hac1Δ cells did not limit growth in glucose-containing medium (Fig. 3F). This contrasts ups1Δ cells and therefore points to additional deficiencies in ups1Δ cells that limit cell growth. We therefore determined transcriptome and proteome profiles of ups1Δ cells under glycolytic growth conditions (Tables S1 and S2). Consistent with the observed impaired UPR activity, canonical UPR target genes were expressed at lower levels in ups1Δ cells when compared with wild-type cells (Fig. S2A). Both transcriptome and proteome profiles pointed to reduced protein synthesis in ups1Δ cells (Fig. 4, A and B). The expression of genes involved in processes driving protein synthesis, such as rRNA processing or ribosome biogenesis, was most prominently downregulated in ups1Δ cells (Fig. 4A and Table S3). Consistently, proteins regulating rRNA processing accumulated at reduced levels in ups1Δ cells (Fig. 4B and Table S4). Together, these data demonstrate that protein synthesis is inhibited in Ups1-deficient cells. The expression of genes related to protein synthesis is decreased under stress conditions (54). Consistently, tunicamycin treatment limited the expression of genes involved in cellular protein synthesis (Fig. 4C). However, it partially promoted cellular protein synthesis in ups1Δ cells (Fig. 4D and Fig. S2C), consistent with the improved cell growth under these conditions (Fig. 3B).
Figure 4.
ups1Δ cells show suppressed ribosome synthesis. A, symplot representing the changed genes within an enriched biological process (BP) GO terms in ups1Δ cells against wild-type (WT) cells. Top ten significant terms from yeast GO-slim are plotted. Percentage of downregulated (blue) and upregulated (red) genes in each term is shown. The size of the central bars was directly proportional to the number of genes in the query belonging to the respective term, and the enrichment value is represented by the color of the bar as previously reported (71). B, 1D annotation enrichment analysis of proteome in ups1Δ cells against WT cells is shown. Annotation is biological process (BP) GO terms (76). Score represents difference of the distribution of the ratios for the proteins corresponding to GO terms from the ratios of the distribution for all proteins. Top ten significant terms are indicated. C, symplot representing the changed genes within a BP GO terms in WT cells treated with 1 μg/ml tunicamycin for 3 h against non-treated WT cells. Top ten significant terms from Yeast GO-slim are plotted. D, symplot representing the changed genes within an enriched BP GO terms in ups1Δ cells treated with 1 μg/ml tunicamycin for 3 h against nontreated ups1Δ cells. Top ten significant terms from yeast GO-slim are plotted. GO, Gene Ontology.
TORC1 inhibition limits glycolytic growth of ups1Δ cells
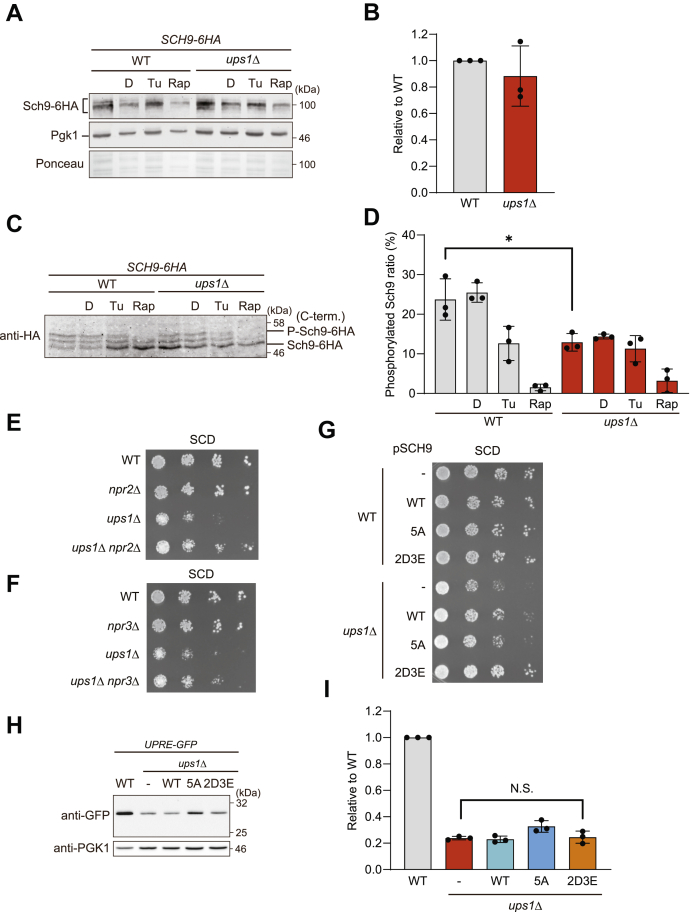
The TORC1 regulates ribosome biogenesis in response to the metabolic status of cells (55). We noted an increased expression of genes coding core autophagy proteins in ups1Δ cells when compared with wild-type cells (Fig. S2B and Table S1), suggesting inhibition of TORC1 and induction of autophagy in ups1Δ cells. To directly assess TORC1 activity in ups1Δ cells, we monitored the level and phosphorylation of one of its substrates, Sch9 (56), an AGC family protein kinase, which is homologous to mammalian S6 and Akt kinases and regulates ribosome biogenesis (57, 58). We found that total Sch9 protein levels were not changed (Fig. 5, A and B), but Sch9 phosphorylation was significantly suppressed in ups1Δ cells compared with wild-type cells (Fig. 5, C and D). These results indicate low TORC1 activity in ups1Δ cells, which agrees with the reduced protein synthesis in these cells.
Figure 5.
TORC1-Sch9 signaling is impaired in ups1Δ cells. A, wild-type (WT) and ups1Δ cells expressing Sch9-6HA grown to log phase in SCD medium were treated with DMSO (D), 1 μg/ml tunicamycin (Tu), or 0.2 μg/ml rapamycin (Rap) for 3 h and subjected to Western blotting. Pgk1 and ponceau staining were monitored as a loading control. B, Sch9-6HA of nontreated cells in (A) was quantified. Total Sch9-6HA signals were normalized to Pgk1 and expressed relative to wild-type cells (set as one). Data are means ± SD (n = 3). C, WT and ups1Δ cells expressing Sch9-6HA grown to log phase in SCD medium were treated with DMSO (D), 1 μg/ml tunicmycin (Tu), or 0.2 μg/ml rapamycin (Rap) for 3 h. For the analysis of Sch9 phosphorylation, lysates were treated with NTCB and subjected to Western blotting. D, Phosphorylated Sch9-6HA ratio in (C) was quantified. Phosphorylated Sch9-6HA signals were divided with total Sch9-6HA signals. Data represent mean ± SD (n = 3). ∗p < 0.05. E, Serial dilutions of WT, npr2Δ, ups1Δ, and ups1Δnpr2Δ cells were spotted on SCD medium and incubated at 30 °C for 2 days (n = 3). F, Serial dilutions of WT, npr3Δ, ups1Δ, and ups1Δnpr3Δ cells were spotted on SCD medium and incubated at 30 °C for 2 days (n = 3). G, Serial dilutions of WT and ups1Δ cells transformed with an empty vector or a plasmid encoding Sch9WT (pSCH9), Sch95A (T723A, S726A, T737A, S758A, S765A), or Sch92D3E (T723D, S726D, T737E, S758E, S765E) were spotted on SCD medium and incubated at 30 °C for 2 days (n = at least 3). H, 4xUPRE-GFP expressing WT and ups1Δ cells transformed with an empty vector or a plasmid encoding Sch9WT, Sch95A, or Sch92D3E were grown to log phase in SCD medium and subjected to Western blotting. I, GFP in (H) was quantified. GFP signals were normalized to Pgk1 and expressed relative to WT cells (set as one). Data represent mean ± SD (n = 3). N.S., not significant; NTCB, 2-nitro-5-thiocyanobenzoic acid; SCD, synthetic complete glucose; UPRE, UPR element.
We hypothesized that, similar to UPR activity, an altered membrane lipid composition may inhibit TORC1 activity in ups1Δ cells. However, neither loss of the CL synthase Crd1 to decrease CL levels (Fig. S3A) nor choline supplementation of wild-type cells to mimic the PE/PC ratio of ups1Δ cells affected TORC1 activity and phosphorylation of Sch9 (Fig. S3, B–E). Similarly, decreased PE levels upon loss of the mitochondrial PS transfer protein Ups2 did not alter TORC1 activity (Fig. S3, B and C). TORC1 activity is dependent on PA (59, 60), but disturbed intramitochondrial PA transport in ups1Δ cells did not result in the accumulation of PA in ER-enriched membrane fractions (Fig. 1, D and E). Moreover, PA accumulation upon inhibition of the PA phosphatase in nem1Δ or spo7Δ cells did not affect Sch9 phosphorylation (Fig. S3, F and G). Thus, other deficiencies appear to inhibit TORC1 activity in ups1Δ cells.
To examine whether TORC1 inhibition limits the glycolytic growth of ups1Δ cells, we used a genetic approach. The Seh1-associated complex inhibiting TORC1 consisting of Npr2, Npr3, and Iml1 functions as negative regulator of TORC1 in yeast (61, 62). Constitutive activation of TORC1 upon deletion of one of the Seh1-associated complex inhibiting TORC1 subunits restored cell growth of ups1Δ cells (Fig. 5, E and F). Moreover, expression of a phosphomimetic mutant variant of Sch9, Sch92D3E, greatly restored the growth of ups1Δ (Fig. 5G). We therefore conclude that inhibition of TORC1-Sch9 signaling impairs the growth of ups1Δ cells on a fermentable carbon source such as glucose. Notably, TORC1 activation upon expression of Sch92D3E in ups1Δ cells did not restore UPR activity (Fig. 5, H and I), indicating that both pathways promote glycolytic growth of ups1Δ cells independently.
Discussion
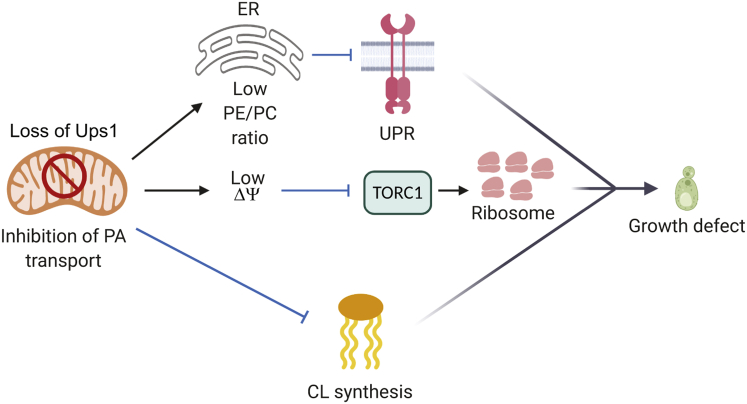
Ups1-dependent transport of PA to the IM promotes CL synthesis (23). Here, we have unraveled an unexpected link between intramitochondrial PA transport and cellular signaling (Fig. 6). Disturbed PA transport in mitochondria lacking Ups1 limits the activation of UPR and inhibits TORC1. TORC1 signaling controls ribosome biogenesis and protein synthesis, which is suppressed in ups1Δ cells and limits glycolytic cell growth. Accordingly, activation of either UPR or TORC1 is sufficient to suppress growth deficiencies in the absence of Ups1. Thus, the flux of PA into mitochondria does not only ensure mitochondrial CL synthesis and membrane biogenesis but is coupled to the phospholipid homeostasis of other cellular membranes affecting cell signaling.
Figure 6.
Altered PA transport in mitochondria affects UPR activation and TORC1 signaling. Impaired PA transport in mitochondria upon loss of Ups1 alters the ER lipid composition (low PE/PC ratio) inhibiting UPR. Independently, TORC1-Sch9 signaling is suppressed in ups1Δ cells. Deficiencies in UPR and TORC1 signaling combined with reduced CL synthesis limit cell growth. CL, cardiolipin; ER, endoplasmic reticulum; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; TORC1, target of rapamycin complex 1; UPR, unfolded protein response.
Several lines of evidences reveal that the disturbed intramitochondrial PA transport in ups1Δ cells impairs independently CL synthesis and UPR and TORC1 signaling. The activation of the UPR allowed glycolytic growth of ups1Δ cells but did not restore the accumulation of CL in mitochondrial membranes (Fig. 3G) or the phosphorylation of Sch9 (Fig. 5, C and D). On the other hand, mimicking the PE/PC ratio observed in ups1Δ cells by choline supplementation of wild-type cells suppressed UPR but did not alter the phosphorylation of Sch9 (Fig. S3, D and E). Conversely, expression of phosphomimetic Sch92D3E mutant did not restore basal UPR in ups1Δ cells (Fig. 5, H and I). These findings are consistent with a previous study demonstrating that activation of TORC1 does not affect the UPR (63). Moreover, CL deficiencies in cells lacking the CL synthase Crd1 did not suppress TORC1 signaling (Fig. S3, B and C) or basal UPR (Fig. S1, E and F). Therefore, we conclude that the impaired intramitochondrial PA transport rather than the CL deficiency leads to the alteration of stress signaling cascades outside of ups1Δ mitochondria.
Inhibition of PA transport across the intermembrane space in ups1Δ cells is associated with an increased PC and PI and a decreased PE/PC ratio but not with a significant accumulation of PA in the ER membrane. This indicates efficient bi-directional transport of PA between the OM and the ER membrane and suggests that PA is rapidly metabolized in the ER, if it is not consumed for CL synthesis in ups1Δ mitochondria. Because PA serves as a common precursor for the synthesis of PC and CL, PA is likely converted to PC resulting in lowered PE/PC ratios in the ER membrane and the inhibition of UPR. Rapid conversion to PC can also explain why impaired PA utilization in mitochondria does not suppress Opi1, the transcriptional repressor of phospholipid synthesis, which is regulated by PA (51). Transcript profiling revealed no increased expression of Opi1 target genes in ups1Δ cells, consistent with the unaltered PA levels and increased PI in ER membranes in ups1Δ cells. The uncontrolled accumulation of PC and PI in the ER membrane may also retard functionality of the ER in addition to the limited UPR activation, contributing to the impaired glycolytic growth in ups1Δ cells.
The low PE/PC ratio in ER membranes suppresses the basal UPR in ups1Δ cells, complementing previous findings which revealed UPR activation upon an increase of the PE/PC ratio (41). The ratio of PE and PC determines membrane packing and curvature stress in the ER membrane, which appears to be sensed by the Ire1 kinase. Indeed, an amphipathic helix adjacent to the transmembrane domain of Ire1 was found to detect biophysical changes at the ER membrane (64). It therefore can be envisioned that the low PE/PC ratio may improve membrane packing and restrain the formation of the X-shaped transmembrane dimer of Ire1.
Protein synthesis is suppressed under stress conditions (54). Our transcriptome data correspondingly show that expression levels of genes involved in ribosome biogenesis are decreased in wild-type cells treated with tunicamycin. On the other hand, tunicamycin induces a number of those genes in ups1Δ cells in accordance with the recovery of glycolytic growth of the cells. One remaining question is how tunicamycin treatment induces genes involved in protein synthesis, although it does not restore TORC1 signaling in ups1Δ cells. In contrast to the previous indication that the suppression of protein synthesis with tunicamycin proceeds independent of the UPR (65), the growth recovery of ups1Δ cells by tunicamycin depends on Ire1 and Hac1. Therefore, our results imply the existence of an UPR-dependent pathway promoting protein synthesis that is induced by proteotoxic stress in ups1Δ cells.
The loss of Ups1 also impairs TORC1 signaling independent of UPR inhibition. TORC1 inhibition in ups1Δ cells is not caused by alterations in the membrane lipid composition of the ER membrane, as neither a decreased PE/PC ratio, nor the accumulation of PA by other means (e.g., inhibition of PA phosphatase complex in nem1Δ or spo7Δ cells; Fig. S3, F and G) resulted in the inhibition of TORC1 activity. Notably, previous studies have reported that TORC1-dependent Sch9 phosphorylation is inhibited by mitochondrial depolarization (66), coupling mitochondrial OXPHOS activity with cellular nutrient signaling. Because mitochondrial membrane potential is low in ups1Δ cells grown in glucose media (35), it is conceivable that mitochondrial depolarization upon inhibition of PA transport interferes with TORC1 signaling in ups1Δ cells. Interestingly, the strong induction of Pdr3, Pdr5, and Pdr15 in ups1Δ cells suggests induction of the mitochondrial compromised protein import response (67), which is in line with mitochondrial depolarization and import defects in ups1Δ cells (35). Therefore, it is tempting to speculate that signaling cascades in response to mitochondrial protein import stress might contribute to the TORC1 inhibition in ups1Δ cells (68). While further studies are needed to elucidate the link between mitochondrial depolarization and TORC1 signaling, our findings highlight that disturbances in the intramitochondrial transport of phospholipids and alterations in the phospholipid composition of mitochondrial membranes can broadly affect cellular signaling.
Experimental procedures
Yeast strains
Saccharomyces cerevisiae strains used in this study are based on S288c or W303. The genotype of the yeast strains is described in the strain list table (Table S5). Genes were deleted by PCR-targeted homologous recombination with the respective forward and reverse primers. Plasmids encoding disruption maker, HphNT1, NatNT2 or KanMX6, were used as template. C-terminal 6xHA tag of Sch9 at its endogenous loci was constructed using PCR-targeted homologous recombination with plasmid encoding 6HA-NatNT2 (69). For construction of cells expressing both Ire1(ΔIII)-3HA-GFP and 4xUPRE-GFP, 4xUPRE-GFP was integrated by PCR-targeted homologous recombination at the URA3 locus in cells expressing Ire1(ΔIII)-3HA-GFP kindly provided by R. Ernst (PZMS, Homburg). 4xUPRE-GFP-URA3 was amplified using as a template genomic DNA of a yeast strain, which contained this gene and which was kindly provided by M. Graef (MPI aging, Cologne) as a template.
Growth conditions
Yeast strains were cultured in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or synthetic complete glucose (SCD) medium (0.17% yeast nitrogen base without amino acids, ammonium sulfate, 0.5% ammonium sulfate, and 2% glucose) supplemented with necessary amino acids at 30 °C. Choline (1 mM; SIGMA) in water, tunicamycin (0.25, 0.5 or 1 μg/ml; Merck) in DMSO or rapamycin (0.2 μg/ml; SIGMA) in DMSO were added to the media when indicated. For cell growth assay, cells were grown in SCD plate for 1 day at 30 °C and suspended in water at a concentration of 0.1 OD600 units. Four microliter of 5-fold serial dilutions of cells were spotted on SCD plates and incubated for 2 days at 30 °C.
Preparation of ER-enriched microsomes
Hundred OD600 units of cells grown to log phase in SCD medium were collected by centrifugation (3000g, 5 min), washed once with H2O, resuspended in Tris-DTT buffer (0.1 M Tris, 10 mM DTT), and incubated for 15 min at 30 °C. Cells were collected by centrifugation (3000g, 5 min), washed once with 1.2 M sorbitol, resuspended in sorbitol-phosphate buffer (20 mM potassium phosphate buffer [pH 7.4], 1.2 M sorbitol) containing lyticase, and incubated for 40 min at 30 °C. Spheroplasts were collected by centrifugation (3000g, 5 min) and resuspended in ice-cold homogenization buffer (0.6 M sorbitol, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 0.3% BSA [fatty acid free], 1 mM PMSF). Whole cell homogenates were subjected to centrifugation (1200g, 5 min, 4 °C). Membrane and microsomes soluble fraction were separated by centrifugation (17,500g, 5 min, 4 °C). Supernatant was transferred to new tubes and subjected to centrifugation (40,000g, 40 min, 4 °C). The pellet was resuspended in 200 μl of pure water, and the protein concentration was determined by BCA assay. Aliquots of 50 μg protein were kept at −80 °C and were subjected to lipid analysis by qMS.
Quantitative mass spectrometry of phospholipids
0.5 OD600 unit of cells or 5 μg protein of ER-enriched microsomes were used for lipid analysis by qMS. Mass-spectrometric analysis of phospholipids was performed essentially as described (70). Briefly, lipids were extracted from samples in the presence of internal standards of major phospholipids (PC 17:0–14:1, PE 17:0–14:1, PI 17:0–14:1, PS 17:0–14:1, PA 17:0–14:1, PG 17:0–14:1 all from Avanti Polar Lipids) and CL (CL mix I, Avanti Polar Lipids LM-6003). Extraction was performed according to Bligh and Dyer with modifications. Final lipid samples were dissolved in 10 mM ammonium acetate in methanol and were sprayed into a QTRAP 6500 triple quadrupole mass spectrometer (SCIEX) by nanoinfusion spray device (TriVersa NanoMate with ESI-Chip type A, Advion). The quadrupoles Q1 and Q3 were operated at unit resolution. Phospholipid analysis was carried out in positive ion mode. PC analysis was carried out by scanning for precursors of m/z 184 at a collision energy (CE) of 37 eV. PE, PI, PS, PG, and PA measurements were performed by scanning for neutral losses of 141, 277, 185, 189, and 115 Da at CEs of 30, 30, 30, 25, and 25 eV, respectively. CL species were identified by scanning for precursors of the masses (m/z 465.4, 467.4, 491.4, 493.4, 495.4, 505.5, 519.5, 521.5, 523.5, 535.5, 547.5, 549.5, 551.5, 573.5, 575.5, 577.5, 579.5, 601.5, 603.5, 605.5, 607.5, 631.5, 715.5, and 771.5 Da) corresponding DAG-H2O fragments as singly charged ions at CEs of 45 eV. Mass spectra were analyzed by the LipidView Software Version 1.2 (SCIEX) for identification, correction of isotopic overlap, and quantification of lipids. Correction of isotopic overlap in CL species was performed using a spreadsheet calculating and subtracting theoretical amounts of [M + 2] and [M + 4] isotopes of each CL species that are isobalic to other CL species. Lipid amounts (pmol) were corrected for response differences between internal standards and endogenous lipids.
Immunoblotting
One OD600 units of cells were collected, washed with H2O, and extracted by alkaline lysis (0.24 M NaOH, 1% 2-mercaptoethanol, 1 mM PMSF). Protein was precipitated in 25% trichloroacetic acid for 10 min on ice. Precipitates were washed with ice-cold acetone two times, dried, resuspended in 50 μl protein sample buffer (60 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 20 mM DTT, 0.025% bromophenol blue), and incubated for 20 min at 40 °C. Samples corresponding to 0.2 OD600 units of cells were separated by SDS-PAGE, immunobloted to nitrocellulose membrane, and immunodecorated with α-GFP (1:5000, ORIGENE), α-PGK1 (1:10,000, Abcam), or α-HA (1:1,000, Roche). Anti-rabbit, anti-mouse, or anti-rat secondary antibodies conjugated to Dylight 800 (1:40,000, LI-COR) were used and detected with the Odyssey Infrared Imaging System (LI-COR). Quantification of the signals was performed using Image Studio Lite (LI-COR).
RNA extraction and quantitative RT-PCR
Total RNA was isolated with the NucleoSpin RNA kit (MACHEREY-NAGEL) from 3 OD600 units of cells, according to the manufacturer's protocol. cDNA was synthesized with 1 μg RNA and the GoScript Reverse Transcription Mix, Oligo(dT) (Promega). Quantitative RT-PCR was performed with Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and the following primers: ACT1 forward 5′-TGTCACCAACTGGGACGATA and reverse 5′-AACCAGCGTAAATTGGAACG; spliced HAC1 forward 5′-GCGTAATCCAGAAGCGCAGT and reverse 5′-GTGATGAAGAAATCATTCAATTCAAATG. QuantStudio 5 (Thermo Fisher Scientific) was used for measurement. Data were analyzed according to the ΔΔCT method normalized to housekeeping gene ACT1.
RNA sequence for transcriptomics
Libraries were prepared using the Illumina TruSeq mRNA stranded sample preparation Kit. Library preparation started with 1 μg total RNA. After poly-A selection (using poly-T oligo-attached magnetic beads), mRNA was purified and fragmented using divalent cations under elevated temperature. The RNA fragments underwent reverse transcription using random primers. This is followed by second strand cDNA synthesis with DNA Polymerase I and RNase H. After end repair and A-tailing, indexing adapters were ligated. The products were then purified and amplified (14 PCR cycles) to create the final cDNA libraries. After library validation and quantification (Agilent tape station), equimolar amounts of library were pooled. The pool was quantified by using the Peqlab KAPA Library Quantification Kit and the Applied Biosystems 7900HT Sequence Detection System. The pool was sequenced with a PE100 run on a NovaSeq6000 sequencer.
Bioinformatics and data analysis for transcriptomics
Raw reads were quantified using the alignment-free quantification tool kallisto version 0.45.0. Reference genome is sacCer3. Gene counts were imported to R version 3.5.1 and normalized to library size using DESeq2 version 1.22.2. General differential gene expression was determined in pairwise comparisons using DESeq2. Functional enrichment of differentially expressed genes was performed using the DAVID API as previously reported (71). The Yeast Gene Ontology-slim list and information of gene function were obtained from the Saccharomyces Genome Database.
Protein lysis and digest for proteomics
One OD600 units of cells were lysed in 40 μl of 2% SDC in 100 mM Tris-HCl [pH 8.0], and lysate was cleared by centrifugation (12,000 rpm, 10 min, 25 °C). Supernatant was subjected to protein concentration determination. In total, 30 μg of protein were used for protein digestion. Proteins were reduced and alkylated by TCEP (10 mM) and CAA (20 mM) for 60 min at 45 °C. 1 μg of LysC endopeptidase was added and incubated at 37 °C or 2 h, followed by addition of 1 μg of Trypsin for digestion at 37 °C for 16 h. Digestion was stopped by addition of TFA to a final concentration of 0.5%. Lysates were cleared (SDC precipitates) by centrifugation, and the supernatant was subjected for desalting using the StageTip (material: SDB-RPS, Affinisep) technique (72).
To generate the peptide spectral library, 2 μl of each sample (all conditions pooled 1:1) was pooled and subjected to high pH reversed phase chromatography. The instrumentation consisted out of a ZirconiumTM Ultra HPLC and a PAL RTC autosampler. The buffer systems consisted out of two buffers. (A) 10 mM ammonium hydroxide and (B) 80% acetonitrile and 10 mM ammonium hydroxide. Peptides were separated according to their hydrophobicity using an in-house packed column (length = 40 cm, inner diameter = 200 μm, 2.7-μm beads, PoroShell, Agilent Technologies) column. The instrument communicated and were controlled using the software Chronos (Axel Semrau GmbH). The gradient length was 60 min, and in total, 12 fractions were collected (1/60 s) and subsequently concentrated using a SpeedVac to complete dryness. Peptides were dissolved in 10 μl 2% formic acid, 2.5% acetonitrile of which 3 μl were injected per liquid chromatography and tandem mass spectrometry run.
Liquid chromatography and tandem mass spectrometry
For the MS/MS spectra library generation, the QExactive HF-x operated in a Top22 data-dependent mode. MS1 spectra were acquired in a mass range of 350 to 1750 m/z, using an AGC target of 3 × 106 and a resolution at 200 m/z of 60,000. MS/MS spectra were acquired at 15,000 resolution using an AGC target of 5 × 105 and a maximal injection time of 22 ms.
For data-independent acquisition measurements, MS1 spectra were acquired using a resolution of 60,000 and an AGC target of 1 × 106. For MS/MS independent spectra acquisition, 48 windows were acquired at an isolation m/z range of 15 Th and the isolation windows overlapped by 1 Th. The isolation center range covered a mass range of 385 to 1043 m/z. Fragmentation spectra were acquired at a resolution of 15,000 at 200 m/z using a maximal injection time of 22 ms and stepped normalized collision energies of 24, 27, 30. The default charge state was set to 4.
Bioinformatics and data analysis for proteomics
The MS/MS data-dependent spectra library was analyzed with MaxQuant 1.6.3.4 and the implemented Andromeda search engine (73) using default settings. The acquired MS2 spectra were correlated against the Uniprot reference Yeast proteome (downloaded 01.2019, 6007 entries). The maximum number of missed cleavages was set to 2. Carbamidomethylation at cysteine residues was defined as a fixed modification, whereas methionine oxidation and protein N-terminal acetylation were defined as variable modification. The false discovery rate (FDR) on the peptide spectrum match and protein level was estimated to 0.01 using the implemented decoy-based algorithm. The mass tolerance on the peptide precursor level to 20 ppm (first search) and 4.5 ppm (main search). On the fragment level (MS2), the mass tolerance was set to 20 ppm. The output txt folder was then used to build the spectral library in Spectronaut (v. 13.0.190604). Data-independent acquisition runs were analyzed using the following settings in Spectronaut (v. 14.10.201222) utilizing the DDA library. Acquired runs were aligned using Precision iRT algorithm in Spectronaut by applying nonlinear regression. The algorithm is based on correlation of extracted ion currents to a consensus elution profile (74). Mass tolerances were set as default (40 ppm for MS1 and MS2 level). Q-value cutoff on precursor and protein level was set to 0.01 using the implemented decoy method “mutated”. A maximum of three precursors were picked per peptide. Quantification was done on the MS2 level using the are under curve. Protein identifications with single peptide identifications in the library were removed as well as proteins that were identified by a single library peptide in DDA runs.
The data were exported in pivot-table format and further processed in Perseus (75). For pairwise comparison a two-sided t test was utilized. The FDR was calculated by a permutation-based approach using 500 permutations and a fudge factor s0 of 0.1 A protein was considered to be significantly differently expressed at a FDR of 5%. No cutoff for the fold change was considered. 1D enrichments (76) were performed in Perseus software using the Gene Ontology annotations of the Uniprot database.
SCH9 phosphorylation assay
2-nitro-5-thiocyanobenzoic acid treatments were performed as previously reported with slight modifications (56). Ten OD600 units of cells were treated with 6% trichloroacetic acid for 5 min on ice, washed twice with ice-cold acetone, and dried using a SpeedVac. The pellets were re-dissolved in 150 μl of urea lysis buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 6 M urea, 1% SDS, 1 mM PMSF, and 1× Protease/Phosphatase inhibitor cocktail (CST)) and lysed with 0.7-mm-diameter zirconia beads in beads beater (3 × 30 s). After incubation for 10 min at 65 °C and centrifugation (20,000g, 2 min), 100 μl of supernatant was transferred to a new 1.5 ml reaction tube. The lysates were mixed with 30 μl of 0.5 M CHES [pH 10.5] and 20 μl of 7.5 M 2-nitro-5-thiocyanobenzoic acid and incubated overnight at room temperature. Each sample was mixed with 50 μl of 4× protein sample buffer (240 mM Tris-HCl [pH 6.8], 8% SDS, 40% glycerol, 80 mM DTT, 0.1% bromophenol blue) and subjected to Western blotting.
Statistical analysis
Quantitative data are presented as arithmetic means ± standard deviation. The statistical significance in related figures was assessed using two-tailed Student's t test. A p values and the number of experiments (n) are described in corresponding figure legend.
Data availability
All data are contained within the manuscript. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (77) with the data set identifier: PXD022321. Transcriptomic data have been deposited to the GEO omnibus with the data set identifier GSE160861.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Martin Graef, Robert Ernst, and Robbie Loewith for the kind gifts of plasmids and yeast strains. We thank the Cologne Center for Genomics for performing RNA sequencing and the MPI aging bioinformatics facility for support in data processing. We also thank other lab members for discussion.
Author contributions
A. E., T. T., and T. L. conceived and designed the research. A. E. performed the majority of experiments and data analysis. M. J. A generated strains and conducted initial analysis and growth assays. H. N. performed proteomic analysis. A. E., T. T., and T. L. wrote the manuscript.
Funding and additional information
This work was supported by JSPS Overseas Research Fellowships to A. E. and by grants of the Deutsche Forschungsgemeinschaft to T. T. (TA1132/2-1) and T. L. (LA918/14-1).
Edited by Dennis Voelker
Footnotes
This article contains supporting information.
Present address for Mari J. Aaltonen: Montreal Neurological Institute, McGill University, Montreal, Quebec, Canada.
Supporting information
Figures S1 to S3; Tables S5 & S6
Table S1
Table S2
Table S3
Table S4
References
- 1.Tatsuta T., Langer T. Intramitochondrial phospholipid trafficking. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:81–89. doi: 10.1016/j.bbalip.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paradies G., Paradies V., De Benedictis V., Ruggiero F.M., Petrosillo G. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta. 2014;1837:408–417. doi: 10.1016/j.bbabio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Musatov A., Sedlak E. Role of cardiolipin in stability of integral membrane proteins. Biochimie. 2017;142:102–111. doi: 10.1016/j.biochi.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Ikon N., Ryan R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017;1859:1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kameoka S., Adachi Y., Okamoto K., Iijima M., Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018;28:67–76. doi: 10.1016/j.tcb.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizzuto M., Pelegrin P. Cardiolipin in immune signaling and cell death. Trends Cell Biol. 2020;30:892–903. doi: 10.1016/j.tcb.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Bertero E., Kutschka I., Maack C., Dudek J. Cardiolipin remodeling in Barth syndrome and other hereditary cardiomyopathies. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165803. doi: 10.1016/j.bbadis.2020.165803. [DOI] [PubMed] [Google Scholar]
- 9.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A., Tyurin V.A., Yanamala N., Shrivastava I.H., Mohammadyani D., Wang K.Z.Q., Zhu J., Klein-Seetharaman J., Balasubramanian K., Amoscato A.A. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2000;2:754–761. doi: 10.1038/35036395. [DOI] [PubMed] [Google Scholar]
- 11.Kuwana T., Mackey M.R., Perkins G., Ellisman M.H., Latterich M., Schneiter R., Green D.R., Newmeyer D.D. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 12.Claypool S.M., Koehler C.M. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 2012;37:32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard A.K., Ortori C.A., Stoger R., Barrett D.A., Chakrabarti L. Mouse mitochondrial lipid composition is defined by age in brain and muscle. Aging. 2017;9:986–998. doi: 10.18632/aging.101204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng K.Y., Watt M.J., Rensen S., Greve J.W., Huynh K., Jayawardana K.S., Meikle P.J., Meex R.C.R. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J. Lipid Res. 2018;59:1977–1986. doi: 10.1194/jlr.M085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke S.L., Bowron A., Gonzalez I.L., Groves S.J., Newbury-Ecob R., Clayton N., Martin R.P., Tsai-Goodman B., Garratt V., Ashworth M., Bowen V.M., McCurdy K.R., Damin M.K., Spencer C.T., Toth M.J. Barth syndrome. Orphanet J. Rare Dis. 2013;8:23. doi: 10.1186/1750-1172-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lev S. Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S.Y., Huang P., Jenkins G.M., Chan D.C., Schiller J., Frohman M.A. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 18.Bektas M., Payne S.G., Liu H., Goparaju S., Milstien S., Spiegel S. A novel acylglycerol kinase that produces lysophosphatidic acid modulates cross talk with EGFR in prostate cancer cells. J. Cell Biol. 2005;169:801–811. doi: 10.1083/jcb.200407123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osman C., Voelker D.R., Langer T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura Y., Kawano S., Endo T. Lipid homeostasis in mitochondria. Biol. Chem. 2020;401:821–833. doi: 10.1515/hsz-2020-0121. [DOI] [PubMed] [Google Scholar]
- 21.Gaigg B., Simbeni R., Hrastnik C., Paltauf F., Daum G. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta. 1995;1234:214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- 22.Kannan M., Lahiri S., Liu L.K., Choudhary V., Prinz W.A. Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER. J. Lipid Res. 2017;58:553–562. doi: 10.1194/jlr.M072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., Langer T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338:815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- 24.Aaltonen M.J., Friedman J.R., Osman C., Salin B., di Rago J.P., Nunnari J., Langer T., Tatsuta T. MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria. J. Cell Biol. 2016;213:525–534. doi: 10.1083/jcb.201602007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyata N., Watanabe Y., Tamura Y., Endo T., Kuge O. Phosphatidylserine transport by Ups2-Mdm35 in respiration-active mitochondria. J. Cell Biol. 2016;214:77–88. doi: 10.1083/jcb.201601082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potting C., Tatsuta T., Konig T., Haag M., Wai T., Aaltonen M.J., Langer T. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 2013;18:287–295. doi: 10.1016/j.cmet.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 27.MacVicar T., Ohba Y., Nolte H., Mayer F.C., Tatsuta T., Sprenger H.G., Lindner B., Zhao Y., Li J., Bruns C., Kruger M., Habich M., Riemer J., Schwarzer R., Pasparakis M. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature. 2019;575:361–365. doi: 10.1038/s41586-019-1738-6. [DOI] [PubMed] [Google Scholar]
- 28.Saita S., Tatsuta T., Lampe P.A., Konig T., Ohba Y., Langer T. PARL partitions the lipid transfer protein STARD7 between the cytosol and mitochondria. EMBO J. 2018;37 doi: 10.15252/embj.201797909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe Y., Tamura Y., Kawano S., Endo T. Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria. Nat. Commun. 2015;6:7922. doi: 10.1038/ncomms8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miliara X., Garnett J.A., Tatsuta T., Abid Ali F., Baldie H., Pérez-Dorado I., Simpson P., Yague E., Langer T., Matthews S. Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes. EMBO Rep. 2015;16:824–835. doi: 10.15252/embr.201540229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu F., He F., Yao H., Wang C., Wang J., Li J., Qi X., Xue H., Ding J., Zhang P. Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex. EMBO Rep. 2015;16:813–823. doi: 10.15252/embr.201540137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miliara X., Tatsuta T., Berry J.L., Rouse S.L., Solak K., Chorev D.S., Wu D., Robinson C.V., Matthews S., Langer T. Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins. Nat. Commun. 2019;10:1130. doi: 10.1038/s41467-019-09089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sesaki H., Dunn C.D., Iijima M., Shepard K.A., Yaffe M.P., Machamer C.E., Jensen R.E. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. J. Cell Biol. 2006;173:651–658. doi: 10.1083/jcb.200603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osman C., Haag M., Potting C., Rodenfels J., Dip P.V., Wieland F.T., Brugger B., Westermann B., Langer T. The genetic interactome of prohibitins: Coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura Y., Endo T., Iijima M., Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 37.Joshi A.S., Thompson M.N., Fei N., Huttemann M., Greenberg M.L. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 2012;287:17589–17597. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao X., van der Veen J.N., Vance J.E., Thiesen A., Vance D.E., Jacobs R.L. Lack of phosphatidylethanolamine N-methyltransferase alters hepatic phospholipid composition and induces endoplasmic reticulum stress. Biochim. Biophys. Acta. 2015;1852:2689–2699. doi: 10.1016/j.bbadis.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., Hotamisligil G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho N., Xu C., Thibault G. From the unfolded protein response to metabolic diseases - lipids under the spotlight. J. Cell Sci. 2018;131 doi: 10.1242/jcs.199307. [DOI] [PubMed] [Google Scholar]
- 41.Thibault G., Shui G., Kim W., McAlister G.C., Ismail N., Gygi S.P., Wenk M.R., Ng D.T. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol. Cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard M.G., Travers K.J., Weissman J.S. Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 43.Walter P., Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 44.Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 45.Cox J.S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 46.Cox J.S., Shamu C.E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 47.Mori K., Ma W., Gething M.J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 48.Credle J.J., Finer-Moore J.S., Papa F.R., Stroud R.M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimata Y., Ishiwata-Kimata Y., Ito T., Hirata A., Suzuki T., Oikawa D., Takeuchi M., Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Promlek T., Ishiwata-Kimata Y., Shido M., Sakuramoto M., Kohno K., Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henry S.A., Kohlwein S.D., Carman G.M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shamu C.E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 53.Miyata N., Goda N., Matsuo K., Hoketsu T., Kuge O. Cooperative function of Fmp30, Mdm31, and Mdm32 in Ups1-independent cardiolipin accumulation in the yeast Saccharomyces cerevisiae. Sci. Rep. 2017;7:16447. doi: 10.1038/s41598-017-16661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broach J.R. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J.R., De Virgilio C., Hall M.N., Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 57.Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P., Tyers M., Rougemont J., Beyer A.L., Loewith R. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30:3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 60.Foster D.A. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab. 2013;24:272–278. doi: 10.1016/j.tem.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dokudovskaya S., Rout M.P. SEA you later alli-GATOR--a dynamic regulator of the TORC1 stress response pathway. J. Cell Sci. 2015;128:2219–2228. doi: 10.1242/jcs.168922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powis K., De Virgilio C. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov. 2016;2:15049. doi: 10.1038/celldisc.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmed K., Carter D.E., Lajoie P. Hyperactive TORC1 sensitizes yeast cells to endoplasmic reticulum stress by compromising cell wall integrity. FEBS Lett. 2019;593:1957–1973. doi: 10.1002/1873-3468.13463. [DOI] [PubMed] [Google Scholar]
- 64.Halbleib K., Pesek K., Covino R., Hofbauer H.F., Wunnicke D., Hanelt I., Hummer G., Ernst R. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell. 2017;67:673–684 e678. doi: 10.1016/j.molcel.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 65.Pincus D., Aranda-Diaz A., Zuleta I.A., Walter P., El-Samad H. Delayed Ras/PKA signaling augments the unfolded protein response. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14800–14805. doi: 10.1073/pnas.1409588111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawai S., Urban J., Piccolis M., Panchaud N., De Virgilio C., Loewith R. Mitochondrial genomic dysfunction causes dephosphorylation of Sch9 in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 2011;10:1367–1369. doi: 10.1128/EC.05157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weidberg H., Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360 doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samluk L., Chroscicki P., Chacinska A. Mitochondrial protein import stress and signaling. Curr. Opin. Physiol. 2018;3:41–48. [Google Scholar]
- 69.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 70.Tatsuta T. Quantitative analysis of glycerophospholipids in mitochondria by mass spectrometry. Methods Mol. Biol. 2017;1567:79–103. doi: 10.1007/978-1-4939-6824-4_7. [DOI] [PubMed] [Google Scholar]
- 71.Metge F., Sehlke R., Boucas J. AGEpy: A python package for computational biology. bioRxiv. 2018 doi: 10.1101/450890. [preprint] [DOI] [Google Scholar]
- 72.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 73.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 74.Bruderer R., Bernhardt O.M., Gandhi T., Miladinović S.M., Cheng L.Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., Reiter L. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 76.Cox J., Mann M. 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13 Suppl 16:S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–d450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S3; Tables S5 & S6
Table S1
Table S2
Table S3
Table S4
Data Availability Statement
All data are contained within the manuscript. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (77) with the data set identifier: PXD022321. Transcriptomic data have been deposited to the GEO omnibus with the data set identifier GSE160861.