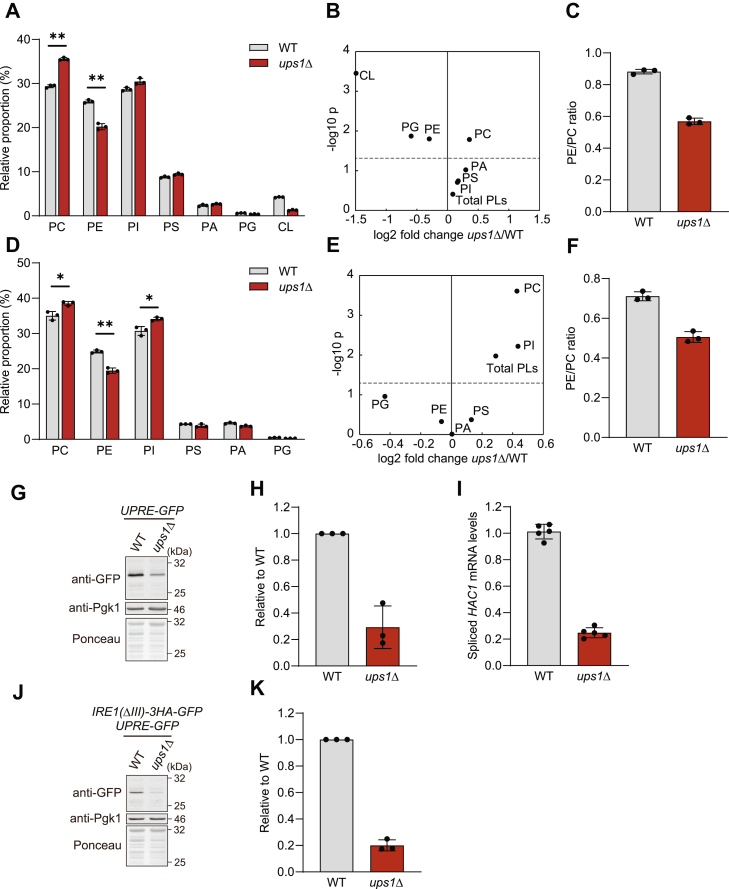
Figure 1.
ER phospholipid composition is altered in ups1Δ cells. A, phospholipid composition in whole cell of wild-type (WT) and ups1Δ cells grown to log phase in SCD medium. Data represent mean ± SD (n = 3). ∗∗p < 0.01. B, changes in the absolute abundance of phospholipids in whole cell of ups1Δ. Data represented as log2 fold change at x-axis with −log 10 p value of student's t test at y-axis. Dashed line represents p = 0.05. C, PE/PC ratio of WT and ups1Δ cells. D, phospholipid composition in ER-enriched microsome fraction from WT and ups1Δ cells grown to log phase in SCD medium. Data represent mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01. E, changes in the absolute abundance of phospholipids in ER-enriched microsome fraction of ups1Δ. Data represented as log2 fold change at x-axis with −log 10 p value of Student's t test at y-axis. Dashed line represents p = 0.05. F, PE/PC ratio in ER-enriched microsome fraction from WT and ups1Δ cells. G, WT and ups1Δ cells expressing 4xUPRE-GFP grown to log phase in SCD medium were subjected to Western blotting. Pgk1 and ponceau staining were monitored as a loading control. H, GFP in (G) was quantified. GFP signals were normalized to Pgk1 and expressed relative to WT cells (set as one). Data represent mean ± SD (n = 3). I, WT and ups1Δ cells were grown to log phase in SCD medium. Spliced HAC1 mRNA expression was analyzed by real time PCR and normalized to ACT1 mRNA expression. Data represent mean ± SD (n = 5). J, WT and ups1Δ cells expressing Ire1(ΔIII)-3HA-GFP and 4xUPRE-GFPgrown to log phase in SCD medium were subjected to Western blotting. K, GFP in (J) was quantified as described in (H). Data represent mean ± SD (n = 3). ER, endoplasmic reticulum; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SCD, synthetic complete glucose; UPRE, UPR element.