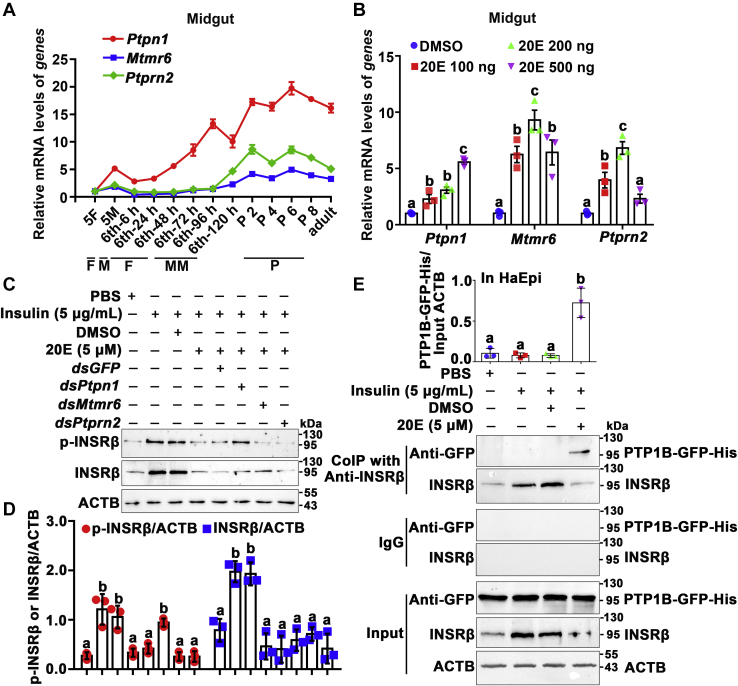
Figure 4.
PTP1B was identified as being involved in 20E-induced dephosphorylation of INSRβ. A, qRT-PCR showing the mRNA expression profiles of Ptpn1, Mtmr6, and Ptprn2 in H. armigera midgut during development based three repeats. B, 20E upregulated the expression of three PTPases in the larval midgut. The sixth instar 6 h larvae were injected different concentrations of 20E for 6 h. Equal volume DMSO was used as a control. All the relative mRNA levels were calculated by 2−ΔΔCT. The bars indicate the mean ± SD of three times repetition. Statistical analysis was conducted using ANOVA, different letters represented significant differences (p < 0.05). C, knockdown of Ptpn1, INSRβ could not be dephosphorylated by 20E induction. The cells were transfected with 4 μg of dsGFP or dsPTPases for 48 h. 20E (5 μM) and insulin (5 μg/ml) were added to the cells for 6 h; 12.5% SDS-PAGE. ACTB as the control. D, statistical analysis of (C) using ANOVA, different letters represented significant differences (p < 0.05). The bars indicate the mean ± SD of three replicates. ImageJ software was used to transform the image data. E, Co-IP showed the interaction between INSRβ and PTP1B-GFP-His. The cells were treated with PBS, insulin (5 μg/ml), insulin plus DMSO, or insulin plus 5 μM 20E for 6 h after transfected with PTP1B-GFP-His for 48 h. The protein expression levels of INSR, PTP1B-GFP-His, and ACTB in HaEpi cells were detected via western blotting following 12.5% SDS-PAGE. INSR was immunoprecipitated with anti-INSRβ, and the coprecipitated PTP1B-GFP-His was detected via western blotting analysis using an anti-GFP mAb. Rabbit IgG was used as negative control of the antibody. Statistical analysis was conducted using ANOVA, different letters represented significant differences (p < 0.05). The bars indicate the mean ± SD of three replicates. ImageJ software was used to transform the image data.