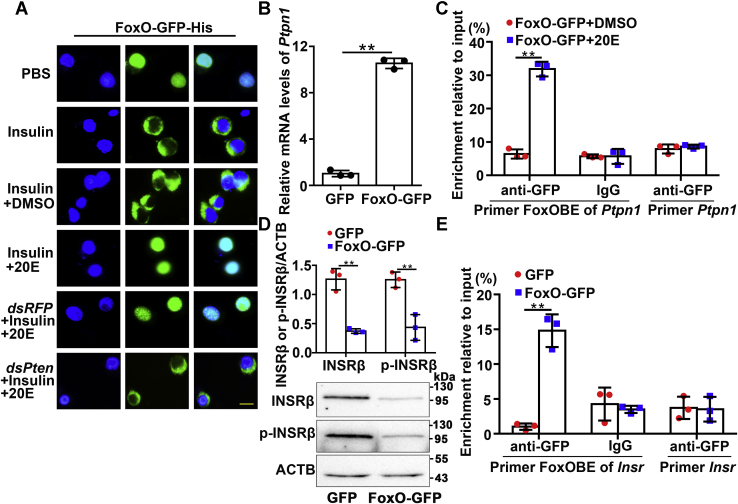
Figure 6.
PTEN determined FoxO nuclear localization and its opposite transcriptional functions in the 20E and insulin pathway.A, the subcellular localization of FoxO-GFP-His in the cells in various conditions (insulin 5 μg/ml, 20E 5 μM for 6 h) without FBS. Green, green fluorescence from FoxO-GFP. Blue, nucleus stained with 4′-6-diamidino-2-phenylindole dihydrochloride (DAPI). Scale bars: 20 μm. B, qRT-PCR showing the increased Ptpn1 expression levels after overexpression of FoxO-GFP in HaEpi cells. C, ChIP analysis of 20E-induced FoxO binding to the FoxOBE in the Ptpn1 promoter region. FoxO-GFP-His was overexpressed in HaEpi cells for 48 h followed by inducing of 5 μM 20E or DMSO for 6 h. Input: nonimmunoprecipitated chromatin. IgG, nonspecific rabbit IgG. Primer FoxOBE of Ptpn1: primers targeted the Ptpn1 FoxOBE-containing sequence. Primer Ptpn1: primers targeted to Ptpn1 ORF. D, western blotting showing the repression of INSRβ expression and phosphorylation by the overexpression of FoxO-GFP; 12.5% SDS-PAGE gel. Antibodies against INSRβ and p-INSRβ were used to detect the INSRβ expression and phosphorylation levels. All statistical analyses of the density of western blotting bands were performed with ImageJ software. E, ChIP analysis of FoxO binding to the FoxOBE in the Insr promoter region. FoxO-GFP-His or GFP-His was overexpressed in HaEpi cells for 48 h. Input: nonimmunoprecipitated chromatin. IgG, nonspecific rabbit IgG. Primer FoxOBE of Insr: primers targeted the Insr FoxOBE-containing sequence. Primer Insr: primers targeted to Insr ORF. All the experiments were performed in triplicate, and statistical analysis was conducted using Student’s t-test (∗∗p < 0.01). The bars indicate mean ± SD.