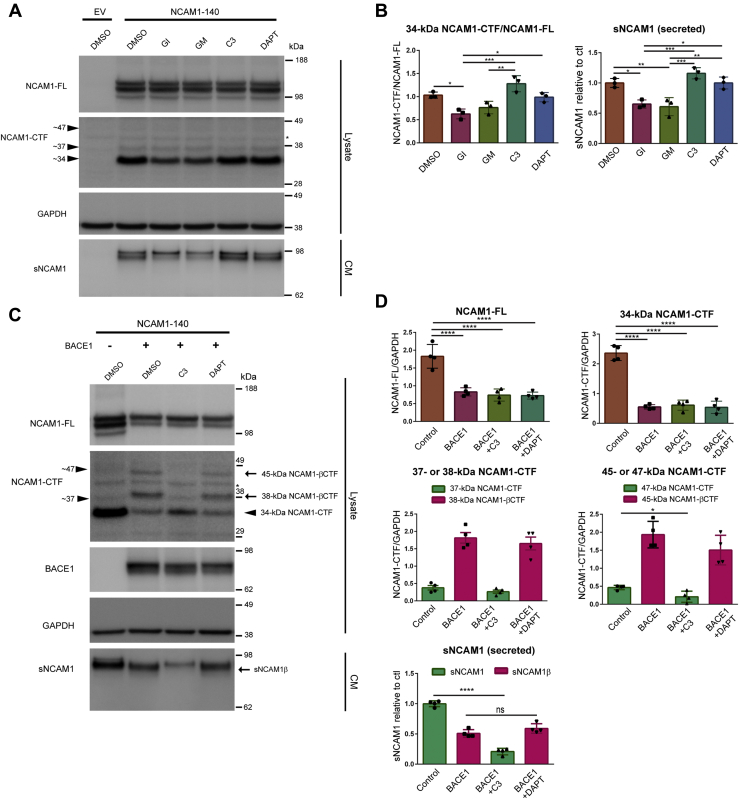
Figure 3.
NCAM1 is cleaved by metalloproteinases or BACE1, but not sequentially cleaved by γ-secretase in HEK cells. HEK cells were transfected with an NCAM1-Myc-DDK expression vector (NCAM1-140 isoform) or an empty vector (EV) as a control and then treated with DMSO only or indicated inhibitors dissolved in DMSO; GI254023X (GI; selective ADAM10 inhibitor, 5 μM), GM6001 (GM; broad spectrum of MMPs inhibitor, 2.5 μM), C3 (inhibitor IV, 10 μM), and DAPT (γ-secretase inhibitor, 10 μM) for 24 h as indicated. Total DNA concentration was kept constant at 1.5 μg for the empty vector. A, representative immunoblot of cell lysates (Lysate) and conditioned media (CM) using anti-Myc (2272), anti-NCAM1 (AF2408), anti-BACE1 (D10E5), and anti-GAPDH (MAB374) antibodies. Ectopically expressed NCAM1 undergoes proteolysis with the production of three C-terminal NCAM1 fragments (NCAM1-CTFs): a major fragment of ∼34 kDa, and two additional fragments of ∼37 kDa, and ∼47 kDa (see arrowheads) and secreted soluble fragments (sNCAM1) in conditioned media. The asterisk denotes nonspecific bands. GI and GM treatments significantly decreased the levels of NCAM1-CTF (∼34 kDa) and sNCAM1, indicating that NCAM1 is cleaved by ADAM10 and MMPs. C, HEK cells were cotransfected with NCAM1 and BACE1 or empty vector as a control and then treated with DMSO, C3, or DAPT for 24 h. Note that the ectopic expression of BACE1 results in cleavage of NCAM1 and the production of BACE1-specific NCAM1-CTF (NCAM1-βCTFs; ∼38 kDa and ∼45 kDa) and soluble NCAM1 (sNCAM1β). However, NCAM1-βCTFs were not accumulated by DAPT treatment, suggesting that NCAM1-βCTFs were not sequentially cleaved by γ-secretase. The asterisk denotes nonspecific bands. B and D, graphs represent densitometric quantification of 34 kDa NCAM1-CTF/NCAM1-FL, NCAM1-FL/GAPDH or NCAM1-CTF/GAPDH ratio in lysates and secreted soluble NCAM1 (sNCAM1) in CM. Note that a different protein ladder was used in this figure compared with Figure 1. One-way ANOVA with Tukey's multiple comparison test was applied. ∗p < 0.05, ∗∗ p <0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, A–D, n = 3. Full-length versions of the western blots in C are shown in Figure S4B. ns, not significant.