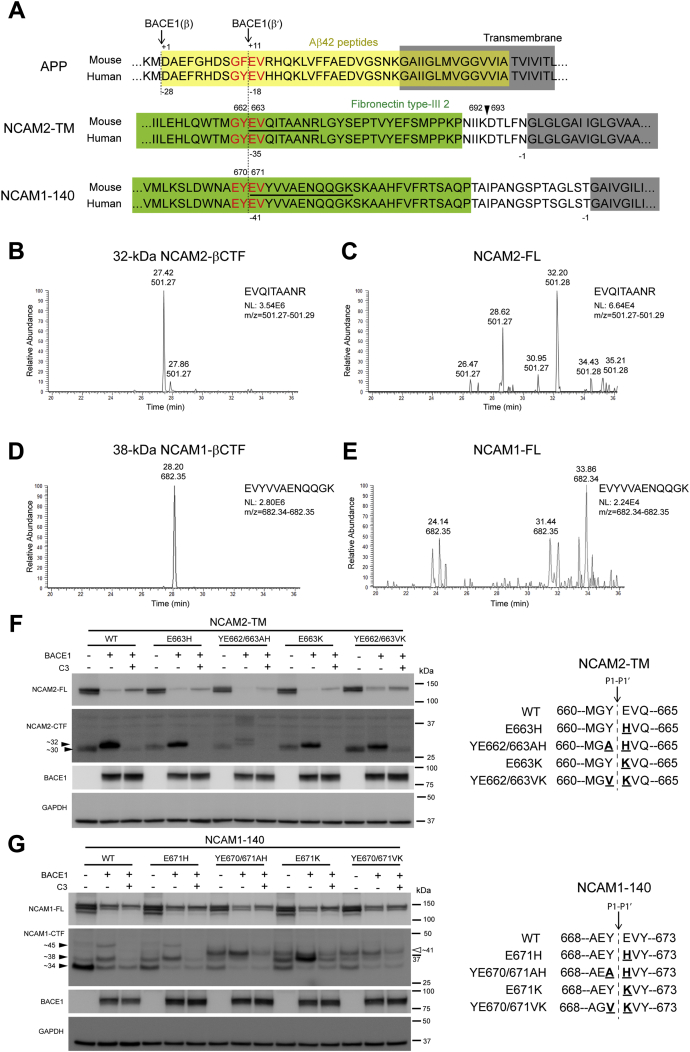
Figure 4.
Identification and validation of the BACE1 cleavage site in NCAM2 and NCAM1. HEK cells were cotransfected with BACE1 and transmembrane NCAM2 (NCAM2-TM) or NCAM1-140. NCAM2-FL and NCAM2-βCTFs; or NCAM1-FL and NCAM1-βCTF were immunoprecipitated in cell lysates using anti-Myc (911B) antibody with agarose beads. After electrophoresis of immunoprecipitated samples, Coomassie stained bands of NCAM2-βCTF (∼32 kDa, in Fig. 2C) and NCAM1-βCTF (∼38 kDa, in Fig. 3C) were cut from the gel and sequenced by microcapillary LC/MS/MS to identify the BACE1 cleavage site. NCAM2-FL and NCAM1-FL were also sequenced as a control. A, comparison of BACE1 cleavage sites on mouse and human APP, NCAM2, and NCAM1. BACE1 cleaves APP at Asp1 within Aβ peptides (yellow box) and also cleaves APP at Glu11 (G680Y681 ↓ E682V683) within Aβ peptides, which are located 28 and 18 residues distant from the transmembrane region (gray box), respectively. APP numbering is based on the 770 amino acid isoform. NCAM2 and NCAM1 numbering is that of the mouse NCAM2-TM and mouse NCAM1-180 amino acid isoform, respectively. The ↓ symbol denotes the scissile (cleavage) bond. BACE1 cleavage sites of NCAM2 (G661Y662 ↓ E663V664) and NCAM1 (E669Y670 ↓ E671V672) in the second fibronectin type-III repeat domain (green box, see FN2 in Fig. 1C) are located 35 and 41 residues distant from the transmembrane region (gray box), respectively. Notably, these cleavage sites are identical or similar to the secondary BACE1 cleavage site (β′) of human APP site (GY↓EV). The cleavage sites of NCAM2 at Asp 693 are indicated by the arrowhead. B and C, ion chromatogram of the peptide peak (EVQITAANR; m/z = 501.27–501.29) from the stained bands (32-kDa NCAM2-βCTF and NCAM2-FL as control). D and E, ion chromatogram of the peptide peak (EVQITAANR; m/z = 682.34–682.35) from the stained bands (38-kDa NCAM1-βCTF and NCAM1-FL as control). F and G, BACE1 cleavage site is denoted by a down arrow depicting the scissile bond (P1-P1′). All of the site-directed mutageneses in P1-P1′ amino acids are denoted. HEK cells were cotransfected with BACE1 and wild-type (WT) NCAM or mutant NCAM and then conditioned for 24 h with solvent only (DMSO) or BACE inhibitor (C3, 10 μM) dissolved in DMSO. Total DNA concentration was kept constant at 1.5 μg with empty vector. Cell lysates were analyzed by immunoblot to assess the BACE1 processing of mutant NCAM2 (E663H, YE662/663AH, E663K, YE662/663VK) (F) or NCAM1 (E671H, YE670/671AH, E671K, YE670/671VK) (G) compared with WT. When BACE1 is overexpressed, only NCAM2-YE662/663AH prohibits the generation of BACE1-specific 32-kDa NCAM2-βCTF (arrowhead). Instead, additional bands are produced, but these bands are abolished by BACE inhibition (C3). These data indicate that BACE1 cleaves NCAM2 between Y662 and E663. NCAM1-E671H mutation did not prevent the generation of NCAM1-βCTFs at ∼38 kDa and ∼45 kDa (arrowhead). The ectopic expression of NCAM1-YE670/671AH, NCAM1-E671K, and NCAM1-YE670/671VK generates additional C-terminal fragments of NCAM1 at ∼41 kDa (41-kDa NCAM1-CTF) compared with NCAM1-WT even in the absence of BACE1 expression (open arrowhead). The coexpression of NCAM1-E671K with BACE1 resulted in increased 38-kDa NCAM1-βCTF levels, concurrently with reduced 45-kDa NCAM1-βCTF levels, most likely by favoring the proteolysis of NCAM1 at Glu 671 by BACE1. In contrast, only double mutations at the P1-P1′ site of NCAM1 (YE670/671AH and YE670/671VK) abolished the generation of BACE1-specfic 38-kDa NCAM1-βCTF and 45-kDa NCAM1-βCTF, but generated 41-kDa NCAM1-CTF, which was reduced by BACE inhibition (C3). These data support that BACE1 cleaves NCAM1 between Y670 and E671. F and G, n = 3.