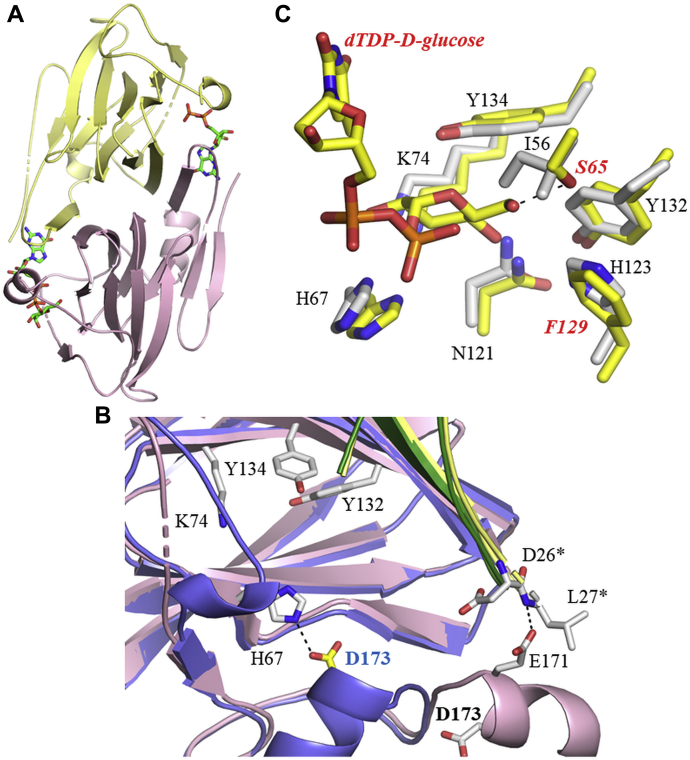
Figure 3.
Structural data on DdahB and MlghB.A, the dimer of DdahB is shown as a cartoon, with one monomer in yellow and the other in pink. GDP and GDP-mannose are shown as sticks with carbons green, oxygen red, nitrogen blue, and phosphorous orange. B, close-up view of the superposition of the DdahB dimer (colored as in panel A) with MlghB (shown in green and blue). Residues shown in sticks are colored with carbons in white for DdahB and in yellow for MlghB. Other atoms are colored as in panel A. Residues numbered with ∗ belong to the second subunit of the dimer. Residues highlighted are those whose role was tested experimentally by site-directed mutagenesis: K74, Y134, Y132 H67, and D173. Asp 173 (shown in bold black for DdahB and blue for MlghB), a key catalytic residue, is out of position in DdahB because of Glu 171 makes hydrogen bond with the other monomer. C, catalytic residues of DdahB monomer are superimposed with Streptococcus suis RmlC (1nyw). The side-chain carbon atoms of DdahB are colored white, whereas carbon atoms of RmlC are colored yellow. The active sites are almost identical, and important residues in DdahB are labeled. Among them, H67, K74 Y134 Y132, and N121 were tested functionally by site-directed mutagenesis. O6 of dTDP-glucose substrate in RmlC hydrogen bonds to Ser 65 and Phe 129 is remote from O6. In DdahB, these residues are found as Ile 56 and His 123. This arrangement is consistent with the larger C7 substrate with C7 making a van der Waal interaction with Ile56 and the hydroxyl on C7 hydrogen bonding to His123.