Fig. 3.
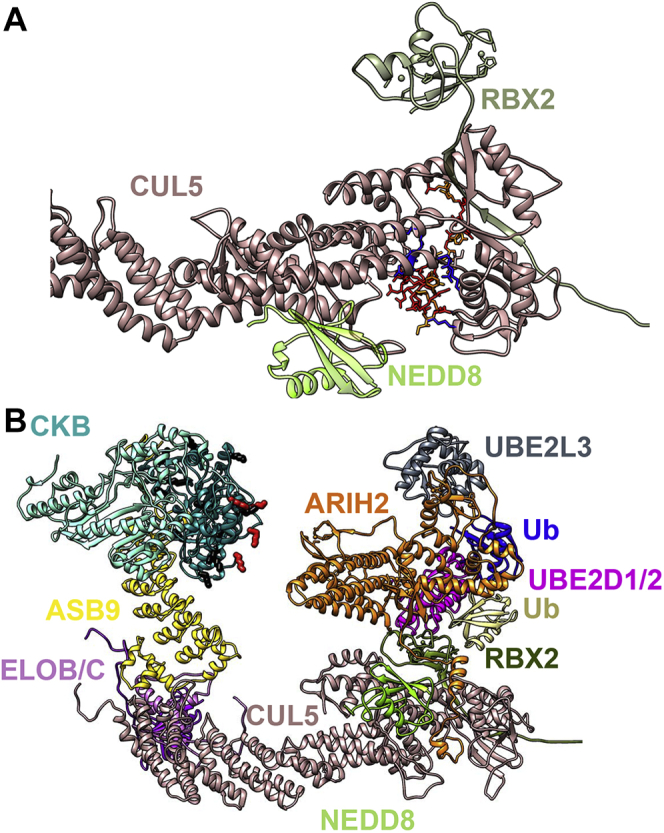
Model of the ASB9-CRL showing sites of ubiquitylation on CKB.A, molecular docking was used to place ARIH2 residues 11 to 38 into the basic cleft in CUL5. Acidic residues on ARIH2 are shown in red and CUL5 basic residues previously shown to be critical for the interaction (21) are shown in blue. B, a structural model of CKB-ASB9-ELOB/C-CUL5(NEDD8)-RBX2-E2D1/2∼Ub-ARIH2-UBE2F∼Ub built from the published structure of ASB9-ELOB/C-CUL5-RBX2-UBE2D1/2 (16) by addition of ARIH2-UBE2F∼Ub based on the docked structure from A). Homologous structural information is not available until residue 57 of ARIH2, so the absolute position of ARIH2 relative to the ASB9-CRL is speculative. The four CKB lysines (K45, K101, K107, and K381) on one subunit of the monomeric CKB that were observed to be ubiquitylated (red side chains) are the closest lysines to ARIH2. The protein colors are consistent throughout the manuscript: CKB, aquamarine, dark cyan; ASB9, yellow; ELOB/C, orchid, purple; CUL5, rosy brown, RBX2, olive; NEDD8, chartreuse; UBE2D1/2, magenta; Ub on UBE2D1/2, khaki; ARIH2, orange, UBE2L3, gray; Ub on UBE2F, blue.
