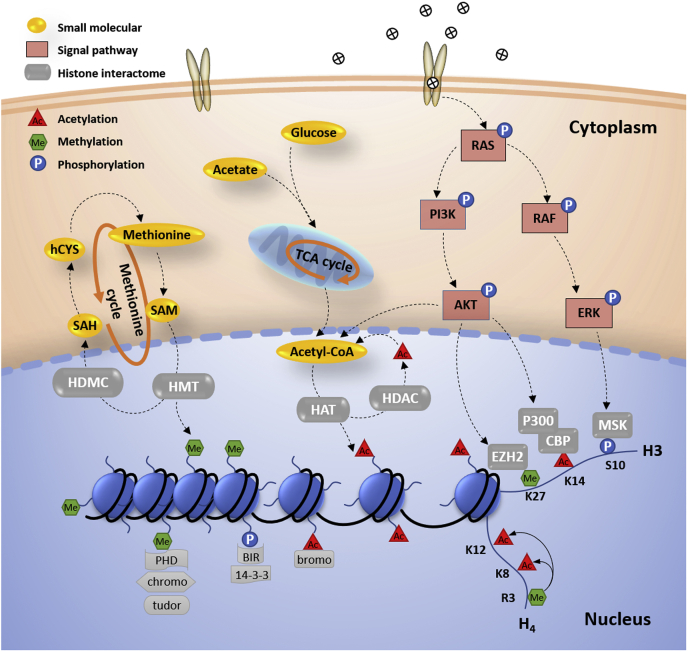
Fig. 1.
Histone PTMs in epigenetic regulation. (I) Two mechanisms by which histone PTMs regulate chromatin functions: (i) direct alternation of chromatin structure, e.g., acetylation leads to an open chromatin; (ii) histone interactome, e.g., plant homeodomain recognizes and binds to methylation and bromodomain to acetylation. (II) Three mechanisms influencing the expression of histone PTMs: (i) histone PTM crosstalk, e.g., the presentence of histone H4R3me promotes the expression of H4K8ac and H4K12ac; (ii) metabolites as the precursors of modifications, e.g., acetyl-CoA/acetylation, SAM/methylation; (iii) signaling pathways triggered by external stimuli, e.g., PI3K/AKT and RAF/ERK modify histone H3 N-terminal tails. AKT, protein kinase B; BIR, baculovirus IAP repeat domain; CBP, CREB-binding protein; ERK, extracellular signal–regulated kinase; EZH2, histone–lysine N-methyltransferase enzyme; HAT, histone acetyltransferase; hCYS, homocysteine; HDAC, histone deacetylase; HDMC, histone demethylase; HMT, histone methyltransferase; MSK, ribosomal protein S6 kinase; PHD, plant homeodomain; RAF, RAF proto-oncogene serine/threonine-protein kinase; RAS, rat sarcoma; SAH, S-adenosylhomocysteine; TCA, tricarboxylic acid.