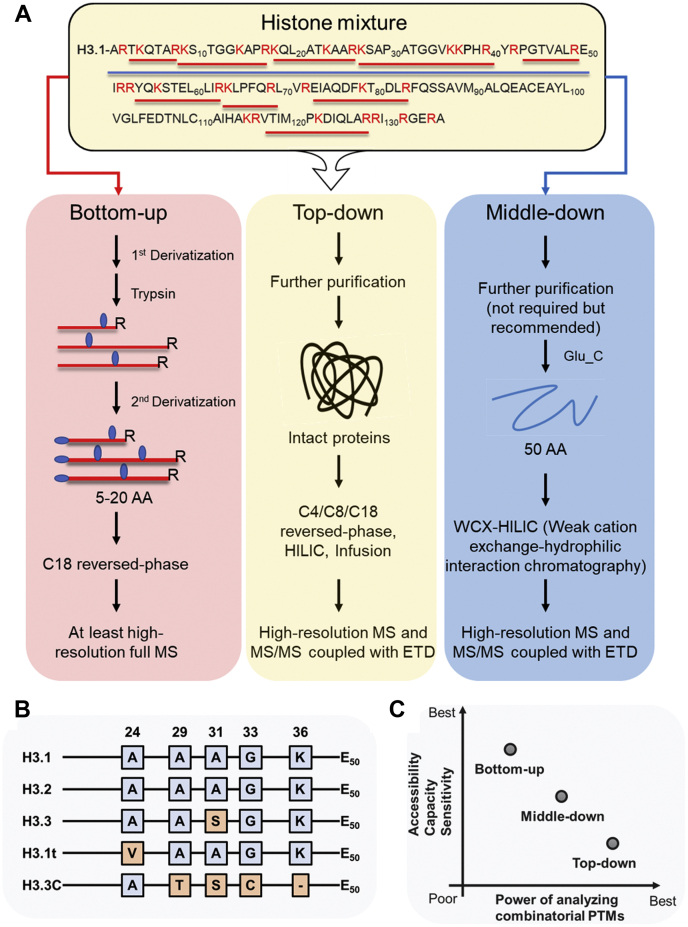
Fig. 3.
MS-based proteomics analysis of histone PTMs.A, the most used workflow for proteomics analysis of histone PTMs (use H3 as example, lysine and arginine residues are labeled in red. Peptides with red underline are sent for bottom–up and blue for middle–down analysis). B, N-terminal tail sequence differences (1–50 AA) between H3 variant representatives. C, comparison of three proteomics approaches in analyzing histone PTMs. ETD, electron-transfer dissociation; MS, mass spectrometry; WCX-HILIC, weak cation exchange-hydrophilic interaction chromatography.