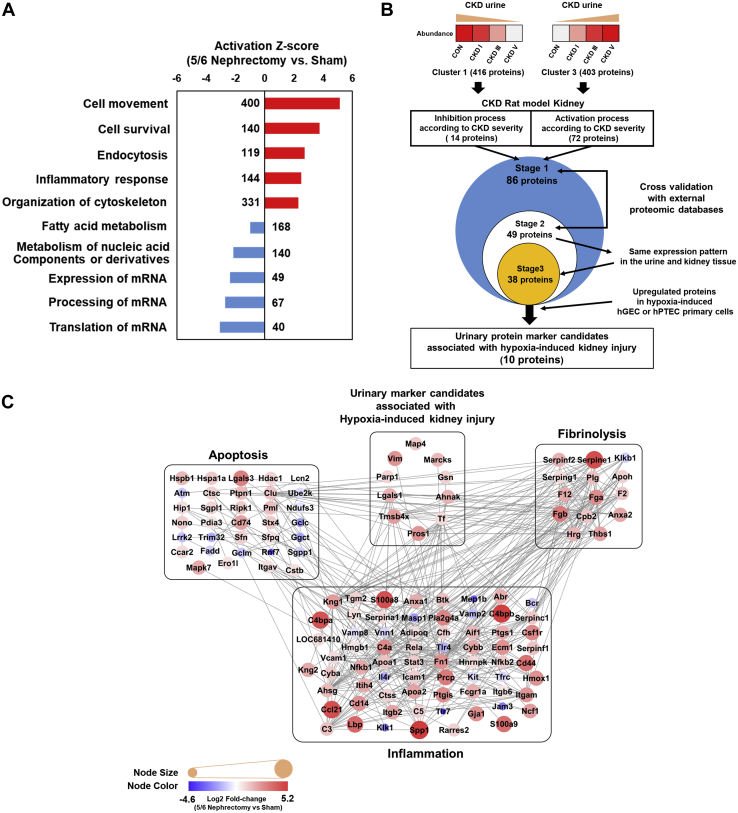
Fig. 5.
Protein identification and protein interaction network associated with chronic injury.A, ten most differentially expressed proteins (DEPs) between nephrectomized and sham-operated groups of chronic kidney disease (CKD). B, schematic flow chart showing sequential analysis along with the number of proteins discovered in each stage of the analysis. Briefly, hierarchical clustered urine DEPs whose levels sequentially increased or decreased with the CKD stages were compared with DEPs identified in CKD rat models to check whether any of these DEPs had same expression patterns in two types of samples (Stage 1). Next, only the significant DEPs were cross validated with external animal experimental data sets (Stage 2) and further shortlisted after comparison of their expression patterns in urine samples (Stage 3). In the final stage, only DEPs that showed same expression patterns in the chronic injury models of human GECs or PTECs were used for further validation analyses. C, protein interaction network model related to inflammation, fibrosis, and apoptosis. The overall organization of the protein–protein interaction was visualized using Cytoscape 3.7.1. Color of the inside nodes indicates the expression levels of quantified proteins. Gray line represents protein–protein interactions obtained from the STRING database. CKD, chronic kidney disease; DEP, differentially expressed protein; GECs, glomerular endothelial cells; FDR, false discovery rate.