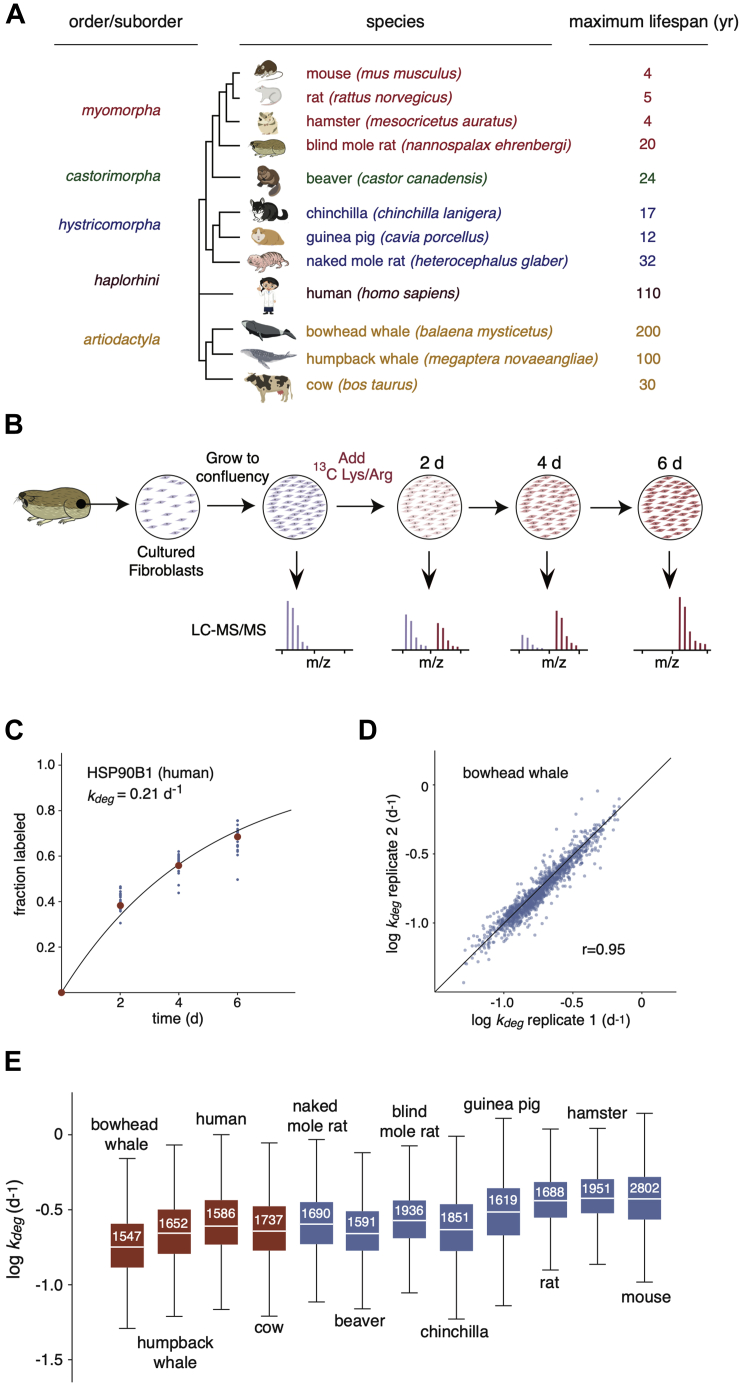
Fig. 1.
Proteome-wide quantitation of kdegin mammalian fibroblasts.A, phylogenetic tree and maximal life spans of species analyzed in this study. Colors indicate distinct orders and suborders. B, dynamic SILAC experimental design. Blue and red colors indicate unlabeled and isotopically labeled spectra/cells, respectively. C, labeling kinetics of human HSP90B1 (endoplasmin) shown as an example of protein-level determination of kdeg values. Blue dots indicate the fraction labeled for all peptides mapped to the protein, and red dots indicate the median fraction labeled for all peptides. The blue curve is a least squares fit to a first-order exponential equation used to measure the indicated kdeg value. D, pairwise comparison of kdeg measurements in biological replicates of bowhead whale cells. The solid line indicates the identity line. The r value indicates the Pearson correlation coefficient. E, distribution of kdeg measurements for each species. The box plots indicate the median (white line), the interquartile range (box), and complete range (whiskers) of kdeg measurements excluding far outliers (>2 SD). The number in each box indicates the number of protein-level kdeg measurements. Red and blue boxes distinguish data collected in this study and the previous study by Swovick et al. (21), respectively. SILAC, stable isotopic labeling in cell culture.