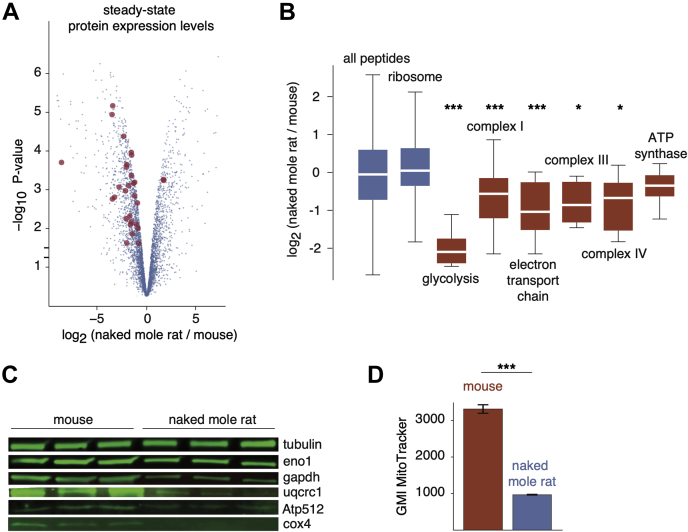
Fig. 5.
Proteome-wide differences in steady-state protein levels between mouse and naked mole rat cells.A, the volcano plot of the p-value versus log2 ratio of expression levels in naked mole rat and mouse cells. Blue points represent all proteins, and red points highlight proteins involved in glycolysis and oxidative phosphorylation with significantly altered expression levels. B, distribution of log2 expression level ratios for specified protein subsets. The box plot representations are as described in Figure 1. Red boxes highlight GO terms involved in ATP production. ∗, and ∗∗∗ indicate p-values of less than 0.05, 0.005, and 0.0005, respectively, in comparison with the global distribution using the Mann–Whitney U test. C, western blots of selected proteins from respiratory chain complexes (Uqcrc1, Atp512, Cox4) and glycolysis (Eno1, Gapdh). D, measurements of geometric mean intensities of MitoTracker mitochondrial staining of naked mole rat and mouse cells. GO, gene ontology.