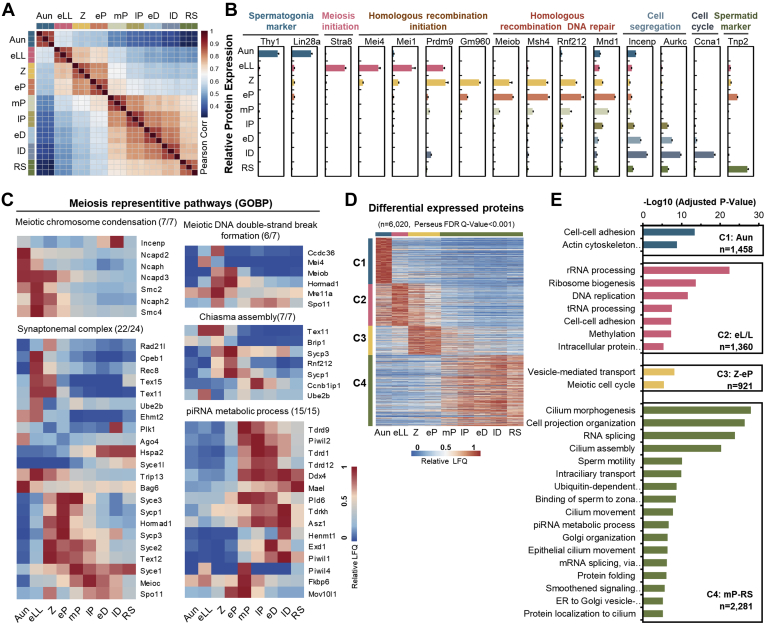
Fig. 3.
Proteomic informatics of the mouse germ cells.A, Pearson correlation coefficients of the log-transformed protein LFQ intensities among the cells at nine substages. The color key represents the value of Pearson correlation coefficients. B, relative abundances of the typical biomarkers in the nine substages of germ cells. Error bars represent SEM in triplicates. C, heat maps of dynamic abundances of the proteins involved in the five representative meiotic pathways. D, K-means clusters of the relative protein abundances elicited from the differentially expressed proteins. The color key represents the relative protein abundances. E, Gene Ontology analysis of the enriched biological processes of the four differentially expressed protein clusters. ER, endoplasmic reticulum; FDR, false discovery rate; GOBP, Gene Ontology Biological Process; LFQ, label-free quantification; piRNA, Piwi-interacting RNA; RS, round spermatid.