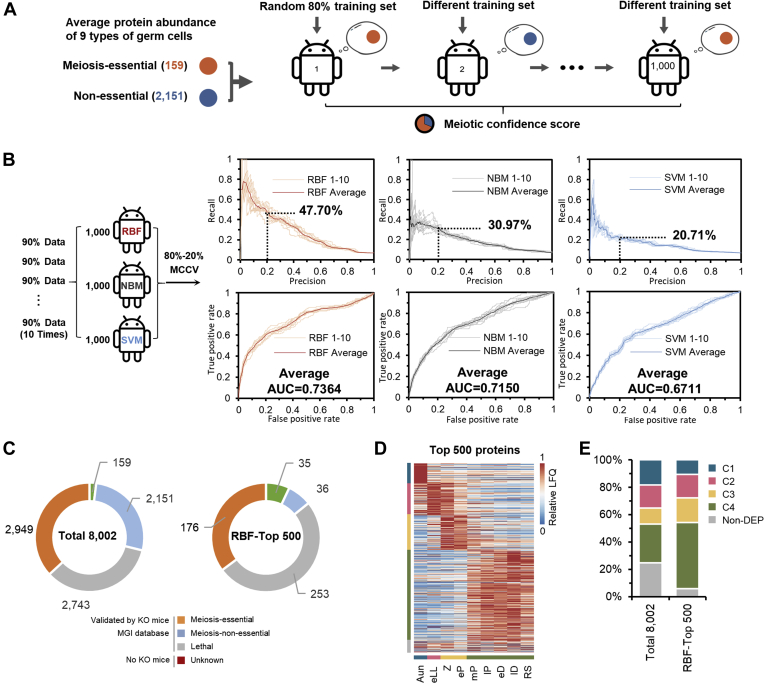
Fig. 5.
Prediction of the meiotic-essential candidates using machine learning. A, workflow to build machine-learning algorithms based on the proteome data.B, comparison of the prediction results, precision-recall curves (upper panels), and receiver operating characteristic curves (lower panels) generated from the three subclassifiers, regularized RBF, NBM, and SVM. The Monte Carlo crossvalidation (MCCV). C, distribution of meiosis-essential, meiosis-nonessential, lethal, and unknown proteins in the 8002 quantified proteins (left panel) and the top 500 meiosis candidates derived from the RBF prediction (right panel). D, heat map of dynamic abundances of the top 500 meiosis candidates derived from the RBF prediction. E, comparison of the distribution of the DEP clusters treated with/without RBF filtration. AUC, area under the curve; DEP, differentially expressed protein; LFQ, label-free quantification; MGI, mouse genomics informatics; NBM, naive Bayesian model; RBF, radial basis function; RS, round spermatid; SVM, support vector machine.