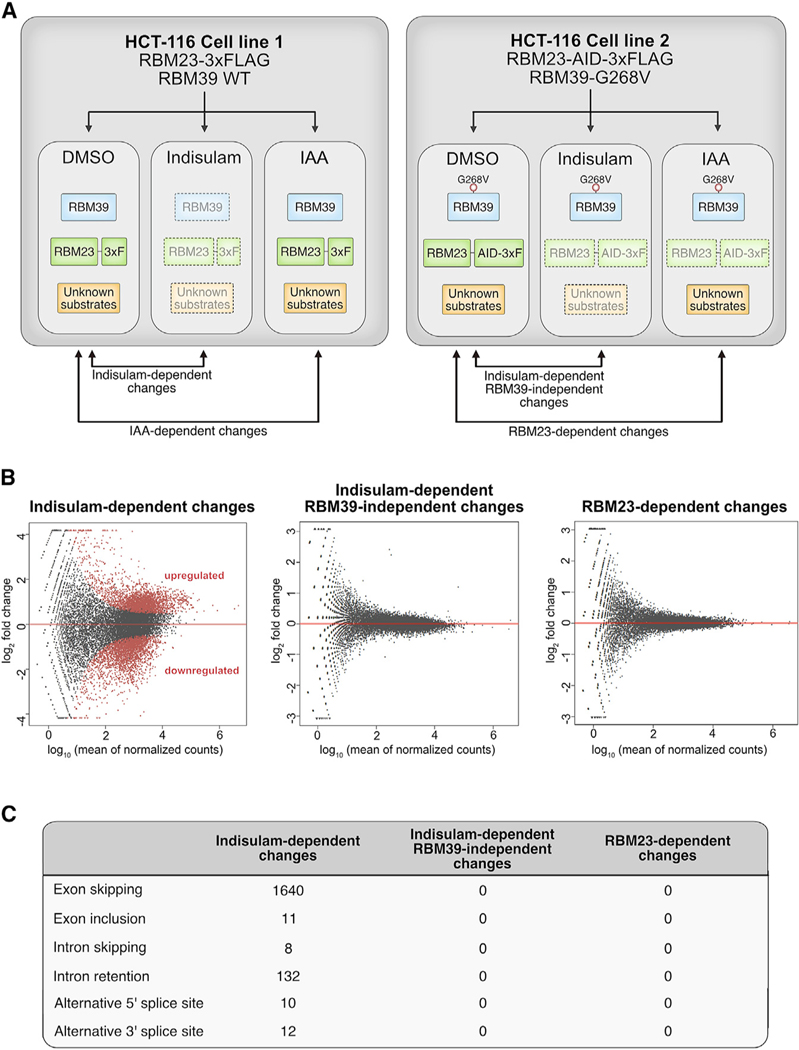
Figure 6. Indisulam-Mediated Gene Expression Is Dependent on RBM39.
(A) Schematic diagram of cell lines used to examine RNA expression and splicing. Endogenously tagged RBM23 cell lines in the background of wild-type RBM39 or RBM39 G268V were treated with DMSO, indisulam (10 μM), or IAA (100 μM) for 12 h. Two clones per cell line were used for gene expression analysis.
(B) MA plots of indisulam-specific, RBM39-specific, and RBM23-specific changes in differential gene expression. Upregulated or downregulated genes with each condition are indicated in red (p < 0.05, log2 fold change > 1). See also Figure S7.
(C) Table of splicing events that are dependent on indisulam, RBM39, or RBM23. Treatment with indisulam, as outlined in Figure 6A, leads to exon skipping and intron retention events that are dependent on RBM39 degradation.