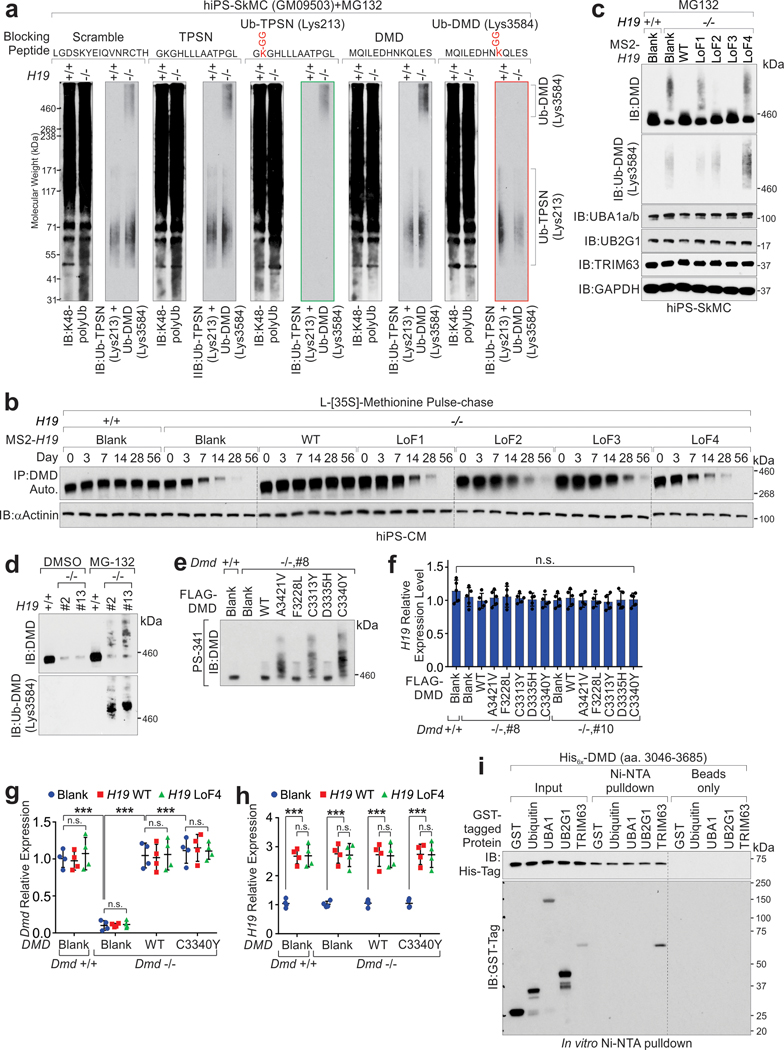
Extended Data Fig. 2. H19 antagonizes poly-ubiquitination and protein degradation of DMD.
a, Blocking Peptide Competition Assay using cell lysates extracted from H19-proficient, or -deficient iPS-SkMCs and blocking peptides of Scramble, TPSN, Ub-TPSN, DMD and Ub-DMD as shown, followed with IB detection using indicated antibodies. MG132 was used as proteasome inhibitor. b, Autoradiography of immunoprecipitated dystrophin in H19-proficient or -deficient iPS-CMs stably expressing MS2-tagged H19 WT or indicated mutants followed by L-[35S]-Methionine pulse-chase. Input was subjected to IB detection using indicated antibody. c, IB detection of indicated proteins in H19-proficient or deficient hiPS-SkMCs expressing MS2-tagged H19 WT or indicated mutants. d, IB detection of indicated proteins in H19-proficient or deficient C2C12 differentiated myotubes with or without MG-132. e, IB detection of DMD in Dmd-proficient or deficient C2C12 differentiated myotubes expressing DMD WT or the indicated mutants with PS-341. f, RT-qPCR detection of H19 relative expression level in Dmd-proficient or -deficient C2C12-differentiated myotubes expressing DMD WT or indicated mutants. Mean values±SD, n=5 independent experiments, one-way ANOVA. g, and h, RT-qPCR detection of Dmd (g) and H19 (h) relative expression level in Dmd-proficient or –deficient C2C12 expressing DMD or H19 WT or indicated mutants. Mean values±SD, n=4 independent experiments, two-way ANOVA. i, IB detection using indicated antibodies of Ni-NTA pulldown using recombinant proteins as indicated. No significance [n.s.], p > 0.05, *, p < 0.05, **, p < 0.01, ***, p < 0.001, and ****, p < 0.0001. Immunoblots are representative of two independent experiments. Statistical source data and unprocessed immunoblots are provided as Source Data Extended Data Fig. 2.