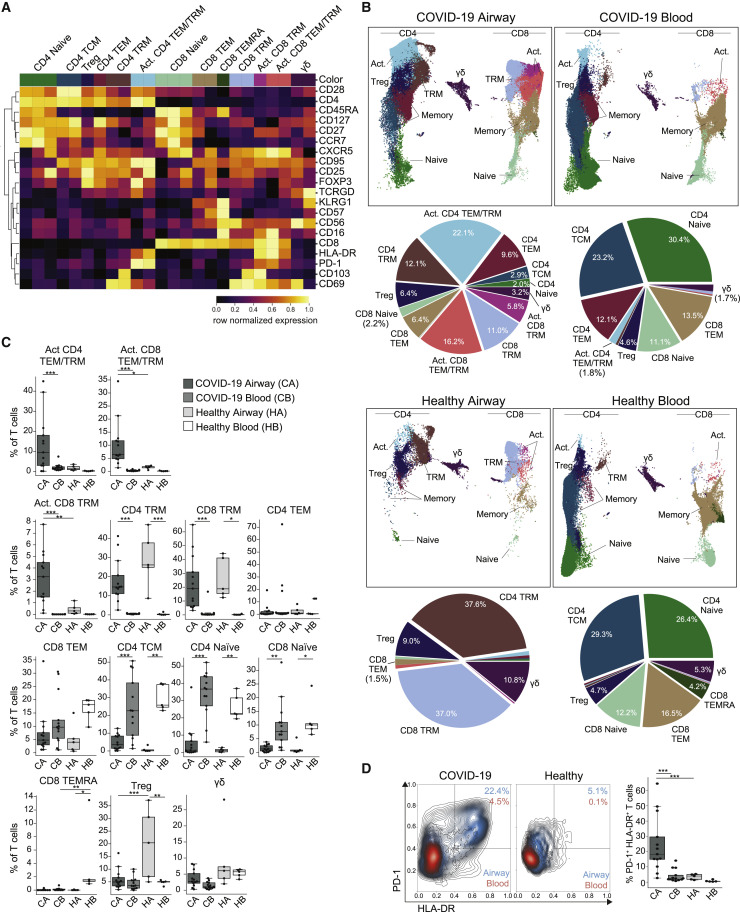
Figure 3.
Airway T cells in COVID-19 are dominated by TRM and activated phenotypes
(A) Heatmap displaying expression of markers within PhenoGraph-generated, hierarchical T cell clusters. A total of 24 PhenoGraph clusters were collapsed into 13 definable T cell subsets indicated along the top. Heatmap data are colored by row normalized value for each marker.
(B) UMAP embeddings of T cell subsets (as defined in A) in the blood and airways of COVID-19 patients and healthy controls (first and third rows). Pie charts indicating relative proportions of defined T cell subsets in airways and blood of COVID-19 patients and healthy controls (second and fourth rows).
(C) T cell subset frequencies in airway and blood samples from COVID-19 patients (n = 13) and healthy controls (n = 5). Boxplots show the frequency of the indicated T cell subset for each patient (average of all time points per patient) or healthy controls. Statistical significance was calculated using a 1-way ANOVA with Tukey’s test for multiple comparisons and indicated by ∗p ≤ 0.05; ∗∗p ≤ 0.01; and ∗∗∗p ≤ 0.001.
(D) Expression of T cell activation markers HLA-DR and PD-1 on T cells (total CD3+ cells) from blood and airways of COVID-19 patients and healthy controls. Left: contour plots showing mean expression on HLA-DR and PD-1 on T cells from each cohort, with airway contours colored in blue and blood colored in red. Right: frequency of T cells expressing HLA-DR and PD-1 in the airways and blood for each COVID-19 patient (n = 13; averaged over all time points) or healthy controls (n = 5). Statistical significance was calculated using a 1-way ANOVA with Tukey’s test for multiple comparisons and indicated by ∗∗∗p ≤ 0.001.