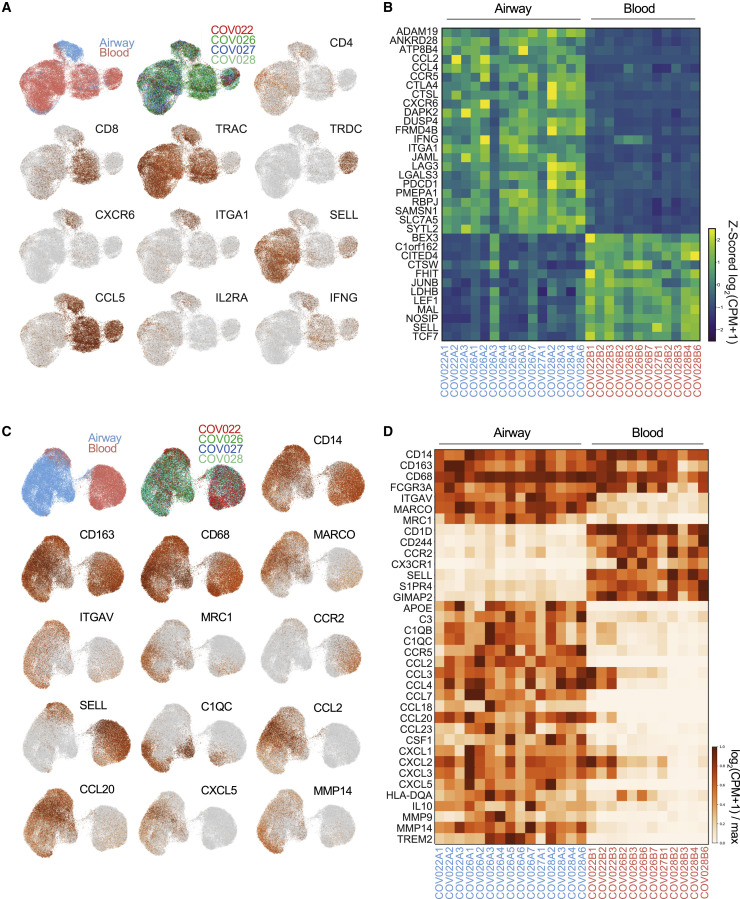
Figure 5.
Inflammatory gene signatures of T cells and myeloid cells are enriched in the airways of COVID-19 patients
T cells, monocytes, and macrophages from the blood and airways of COVID-19 patients were analyzed by scRNA-seq (see Method details).
(A) Separate UMAP embeddings showing gene expression from total T cells obtained from airways and blood of paired samples from 4 patients. UMAP embeddings show sample site origin, subject, and indicated gene expression (based on log2(CPM+1)).
(B) Heatmap showing major differentially expressed genes in airway compared to blood T cells from each individual patient and time point. Data are colored by row Z scored log2(CPM+1) for each sample.
(C) UMAP embeddings of total monocytes and macrophages obtained from airways and blood from 4 COVID-19 patients. UMAP embeddings show sample site origin, patient, and selected gene expression displayed as log2(CPM+1)/maximum.
(D) Heatmap of subset-defining genes, homing receptors, and key inflammatory molecules for monocytes and macrophages in airways and blood from each patient sample. The heatmap shows genes that are not differentially expressed between airways and blood (CD14-FCGR3A) and genes are consistently differentially expressed (ITGAV-TREM2).