Figure 2. Conditional Kmt2c deletion expands the HSC pool at the expense of HPCs.
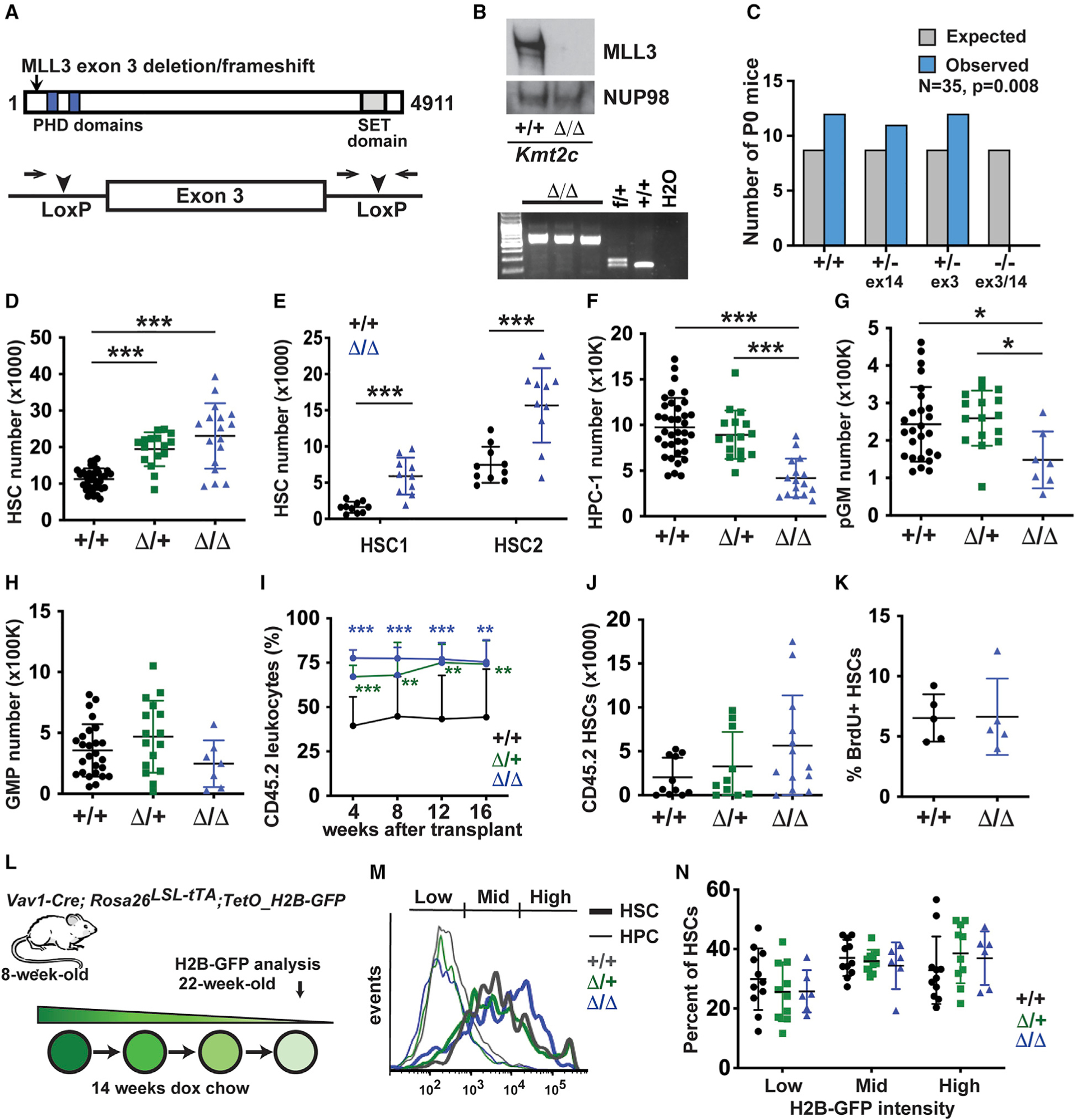
(A) Overview of the conditional Kmt2c-flox allele.
(B) Western blot and genotyping PCR showing loss of MLL3 protein expression in Kmt2cΔ/Δsplenocytes.
(C) Complementation testing of the Kmt2c null (exon 14) and Kmt2cflox (exon 3) alleles, as measured by viable offspring after germline Kmt2cflox deletion. Observed and expected allele frequencies were compared by the chi-square test. n = 35.
(D–H) HSC, HPC-1, pGM, and GMP numbers in 8-week-old mice of the indicated Kmt2c genotypes (two hindlimbs). n = 7–20.
(I) Peripheral blood CD45.2+ leukocyte chimerism after competitive whole bone marrow transplants. n = 10–13 per genotype from three independent donors.
(J) CD45.2+ HSC numbers in primary recipient bone marrow 16 weeks after transplantation.
(K) Twenty-four-hour BrdU incorporation in wild-type and Kmt2cΔ/Δ HSCs. n = 5.
(L) Overview of H2B-GFP pulse-chase experiment.
(M) Representative histograms showing H2B-GFP signal in HSCs and HPCs after 14 weeks of doxycycline exposure.
(N) Percentages of H2B-GFPHigh, H2B-GFPMid, and H2B-GFPLow HSCs after a 14-week doxycycline chase. n = 6–11.
In all panels, error bars reflect standard deviation. *p < 0.05, **p < 0.01, ***p < 0.001; comparisons were made by two-tailed Student’s t test or one-way ANOVA with Holm-Sidak post hoc test (multiple comparisons). See also Figures S1 and S3.
